Research Article - (2022) Volume 10, Issue 8
Testing for SARS-COV-2: Molecular and Serological Diagnosis
Pranjali Jain* and Swarupa Chakole
*Correspondence: Pranjali Jain, Department of Community Medicine Jawaharlal Nehru Medical College, Datta Meghe Institute of Medical Sciences (DU), Sawangi Meghe, Wardha, Maharashtra, India, Email:
Abstract
The coronavirus 2 (SARS-CoV-2) epidemics and accompanying illness (COVID-19) have reached global proportions, putting additional strain on healthcare systems to contain and manage COVID-19. To curb the spread of virus, improvements in diagnosis is needed along with mass screening of a large proportion of population. Search for viral RNA in upper and lower respiratory airways using RT-PCR methods are on-going which will be used to make a molecular diagnosis of SARS-CoV-2 infection and illness. Serological testing for viral antibodies could aid healthcare personnel in supporting COVID-19 diagnosis and follow-up, as well as population screening. First step is to collect the specimen as early as patient come. Site of collection of swabs should be anatomically appropriate for accurate result of test. Main diagnostic test is molecular test that is RTPCR in initial stage whereas supportive test is serological test. CoV-2 is made up of single-strand ribonucleic acid (ssRNA) along with a positive sense strand which is enveloped. Patients with comorbid diseases such as diabetes, cancer, hypertension, malnutrition, old age, children, and pregnancy have severe infection and a high mortality rate. Most appropriate and frequently used test for diagnosing covid-19 is RTPCR. It makes use of TaqMan fluorogenic probe-based chemistry and Taq DNA polymerase's 5′-nuclease activity. Early discovery, diagnosis, isolation, and treatment are the most efficient methods for prevention and control of COVID-19. Both clinical and preclinical knowledge is important for fighting COVID-19 like pathology, microbiology and other detection techniques required for molecular diagnosis and improving ability of early diagnosis.Keywords
SARS-CoV-2, COVID-19, Molecular diagnosis, Serological diagnosisIntroduction
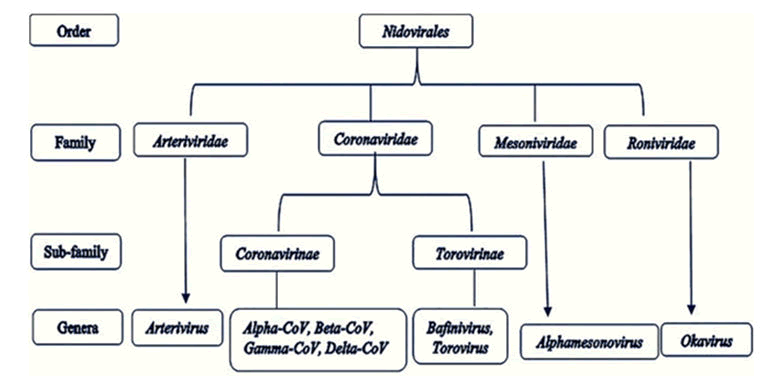
Since December 2019, humanity has been dealing with a pandemic produced by the SARS-CoV-2 beta coronavirus. Coronavirus illness 2019 was named after the disease induced by SARS-COV-2 infection (COVID-19). CoV is a member of the Corona virinae subfamily of the Corona viridae family in the order Nidovirales. Alpha coronavirus, Beta coronavirus, Gamma coronavirus, and Delta coronavirus are the four genera that make up this subfamily. The genome of the virus is a single-stranded positive-sense RNA (+ssRNA) with a 5′ cap structure and a 3′ poly-A tail (approx. 30 kb in size). A normal CoV genome and sub genomes may have 6, or possibly more, open reading frames (Figure 1) [1].
Figure 1: Order nidovirales: taxonomy.
How SARS-CoV-2 is transmitted is unknown, however it is probably similar to SARS, which is disseminated by droplets, contacts and aerosol, and contaminated environments and items. Patients with flu like symptoms that is there can be involvement of respiratory system have a higher risk of transmission, but it can also happen to asymptomatic patients, according to studies. The virus primarily affects the human respiratory system, but follow-up of COVID-19 patients has revealed that the virus can cause symptoms other than respiratory, as well as inflammatory complications in several organ systems, including the gastrointestinal tract, hepatobiliary system, and post-COVID neurological complications. Direct molecular diagnostic testing based on SARS-CoV-2 sequencing is proved to be essential in identifying affected people [1]. In view of restricting extent of coronavirus epidemics, the diagnostic sensitivity and specificity of suspected COVID-19 infection, it is necessary to base decisions on basis of controlled testing and performance data from clinical context. Patient health may be at risk if unreliable and unproven diagnostics overlook present infections or produce misleading positive and negative results (Figures 2 and 3).
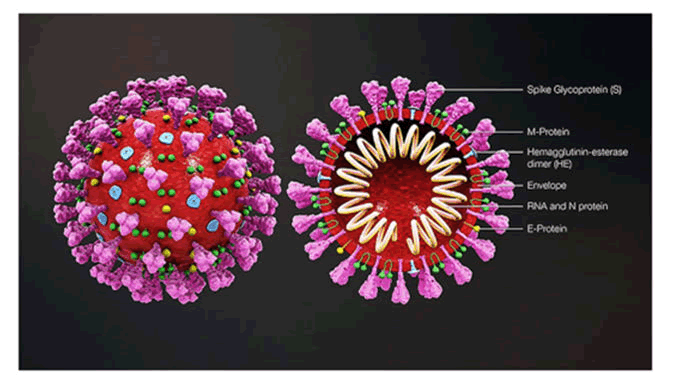
Figure 2: 3D COVID-19 virus structure.
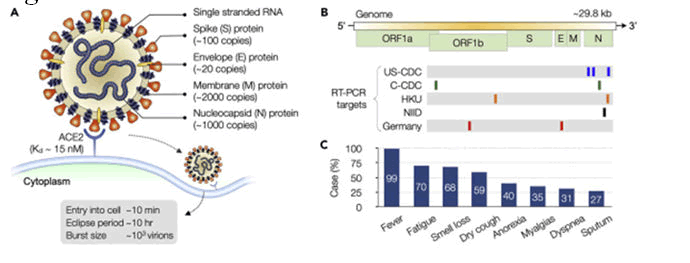
Figure 3: Virus structure and symptoms.
Corona virus is encapsulated and spherical in shape (about 120 nm in diameter), consisting of pointed spikes on surface which are about 20 nm in length. The spike (S protein approx. 100 copies), envelope (E protein approx. 20 copies), and membrane (M protein approx. 2000 copies) are the structural proteins which are attached with the covering that is viral envelop. Inside the envelop, the nucleocapsid create a helical nucleocapsid with the genomic RNA [2].
Single strand RNA genome of virus is organised as follows
5' strand-UTR (untranslated region) replicase S-protein, E-protein, M-protein, N-protein UTR poly (A) tail ending on 3’ is the genome's organisation. Replicase gene is coded by Open Reading Frames (ORFs) 1a and 1b. RT-PCR specific genes for COVID-19 are US-CDC, C-CDC, HKU, NIID, and Germany. N gene has low Sequence conservation therefore several assays target it [2].
Patients have symptoms that are similar to the influenza and cold, while some positive cases are asymptomatic. Highly specific molecular testing must be used to confirm diagnosis [2].
Clinical features: Corona virus causes number of symptoms; it could be a patient without any symptoms to a patient going into shock and multiple organ failure syndromes. COVID-19 is graded according to the severity of the symptoms. Disease is divided into 4 categories:
- Mild,
- Moderate,
- Severe,
- And critical.
Fever (98%), tiredness (70%), cough, and diarrhoea are most prevalent symptoms among patients [3].
Mild disease: Symptoms of an upper respiratory tract infection may be present with a mild disease example cough, cold, fever, body ache, headache and loss of smell. It can be differentiated from severe disease by lack of significant manifestations example: difficulty in breathing. The majority of cases (81%) are less severe. In such circumstances, radiographic characteristics are also missing. Patients with a minor condition might suddenly progress into severe or life-threatening situations [3].
Moderate disease: Cough, shortness of breath, and tachypnoea are common respiratory symptoms in these patients. However, there are no indications or symptoms of a serious illness [3].
Severe disease: Severe pneumonia is seen in patients with serious illness. Respiratory distress/sepsis/or septic shock is seen in patients with excessive pneumonia. Clinical diagnosis is mainstay and radiographic investigations may be used to rule out complications. Within 24 to 48 hours, severe difficulty in breathing, increased breathing rate greater than 30 cps, respiratory distress, partial pressure of oxygen of about 93 percent, Partial pressure of oxygen/Fraction of inspired Oxygen 300 or more than 50 percent lung infiltrates are common clinical features [3].
Respiratory distress syndrome: The beginning of Respiratory Distress shows newly developed or worsened respiratory insufficiency. PaO2/FiO2 measurements are used to distinguish distress on the basis of grades of hypoxia. A Partial pressure of oxygen/Fraction of inspired Oxygen ratio of less than 100 mm Hg indicates extreme ARDS.
PaO2/FiO2 levels between 100 mm of Hg to 200 mm of Hg are signifies moderate ARDS. PaO2/FiO2 levels between 200 mm of Hg and 300 mm of Hg are suggestive of mild ARDS.
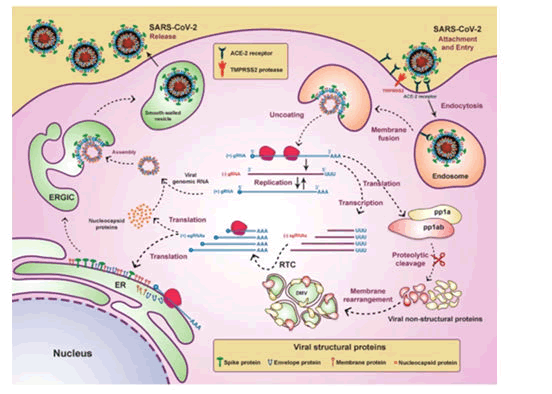
Worsening of disease is linked to Aspartate transaminase and Alanine transaminase level at time of admission and in future. As a result, higher levels at the time of admission lead to a rapid clinical deterioration towards ARDS [3]. Septic Shock and sepsis: COVID-19 and sepsis can be considered the most dangerous combination. Due to downregulated host response to COVID-19 infection, multi organ failure syndrome develops. serious dyspnoea, very low oxygen saturation, decreased urine volume, increased heart rate, decreased blood pressure, cold extremities like in shock, skin wrinkling, and neurological deficit are all signs of organ dysfunction. Acidosis, increased lactate, increased bilirubin content in blood, decreased platelet counts, and indications of coagulopathy are examples of laboratory evidence of additional homeostatic dysregulation (Figure 4) [3].
Figure 4: Life cycle of CoV-2 in host cell.
The virus reaches the cytoplasm via the endosomal route after binding to its receptor. Genomic RNA translates to form viral replicase gene which leads to formation of polyproteins
- Pp 1a
- Pp 1ab
Polyproteins (pp 1a and pp 1ab) produce a variety of other proteins which are known as non-structural proteins. All non-structural proteins combine to form a complex. This complex provide a suitable region for viral RNA which is further responsible for multiplication and transcription of sub genomic RNA and the complex is known as RTC that is Replicase Transcriptase complex. Sub genomic RNA (sgRNA) play a role of mRNAs for structural and other genes present in downstream of replicase. Genomic RNA and sub genomic RNA is made with process of frameshifting (ribosomal) which produce intermediate RNA. S protein, E protein, and M protein that are the structural proteins of viral RNA translates in Endoplasmic Reticulum (ER) and they move in endoplasmic Reticulum-Golgi intermediate compartment ERGIC. Virus genome, which has its protection of N protein, then goes inside ERGIC’s membrane consisting of other structural proteins of virus. Virus particles are then arranged and transferred outside of cell via vesicles, where exocytosis occurs [4].
Sample collection
Specimens are to be collected from:
Upper respiratory tract: Naso Pharyngeal (NP) and Oro Pharyngeal (OP) swabs.
Lower respiratory tract: Aspirates, Broncho Alveolar Lavage (BAL), and nasopharyngeal wash/aspirate.
Alternative sources include saliva, anal swabs, urine and stool, tears, and conjunctival secretions. Sample must be collected and kept in sterile tube already half-filled by Transport Medium (VTM) and placed in refrigerator (2-8°C) for 3 days after collection for travel and storage. Swab should be kept at 70°C or lower when there is delay in processing, transporting and testing of sample [2]. Sample should be shipped in a three-package arrangement. Every vial should be correctly sealed and labelled, and retained:
- Outer absorbent material covering (which is primary or first container) before being placed in the secondary or second container.
- Secondary or second container should be kept in the outer container with frozen gel packs.
- Every step should follow WHO guidelines (Figure 5).
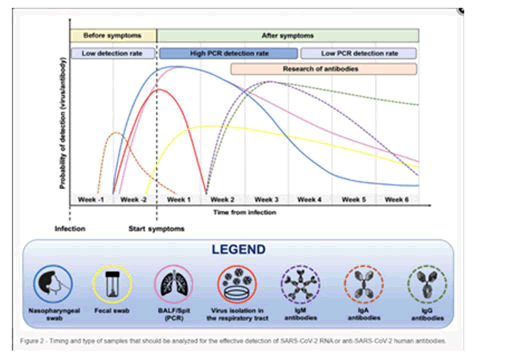
Figure 5: When should SARS-CoV-2 RNA or anti-SARS-CoV-2 human antibodies be tested, and what kind of samples should be tested.
Diagnosis
The pathogenic organism must be identified before an infectious disease may be diagnosed. Virus isolation and viral RNA detection are the most used methods for identifying pathogenic organisms. The majority of COVID-19 diagnosis is nucleic acid testing which are based on viral RNA detection. Viral load in nasopharyngeal, oropharyngeal and sputum samples of extremely diseased patients is higher than mildly diseased patients, according to a study that looked at the viral RNA content of novel coronavirus in 90 infected patients. Eleven extremely diseased patients have more nucleic acid content of viral RNA in sample of sputum when compared with mild diseased patients [5]. Recent evidence is suggestive of COVID infection is linked to conjunctivitis, dermatologic symptoms (maculopapular and vesicular lesions), and Multisystem Inflammatory Syndrome in Children (MIS-C), which is clinically similar to Kawasaki illness. Acute strokes and myocardial infarctions have also been observed, implying multi-organ involvement, which is being investigated further in a number of researches. The Israeli Ministry of Health has developed a clinical prediction model that is independent of RT-PCR and is based on eight factors (cough, fever, contact with a confirmed case, gender, age 60+, headache, sore throat, and shortness of breath) [6].
Materials and Methods
Based on how viral RNA is processed and detected, there are three major molecular methods which are: Real-Time Reverse Transcription Polymerase Chain Reaction (rRT-PCR), isothermal ampliï¬cation, and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) based methods. All these methods follow the same protocol that has been recommended by the Centres. Based on how viral RNA is processed and detected, there are three major molecular methods which are: Real-Time Reverse Transcription Polymerase Chain Reaction (rRT-PCR), isothermal ampliï¬cation, and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) based methods. All these methods follow the same protocol that has been recommended by the Centres.
Based on how viral RNA is processed and detected, there are three major molecular methods which are: Real-Time Reverse Transcription Polymerase Chain Reaction (rRT-PCR), isothermal ampliï¬cation, and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) based methods. All these methods follow the same protocol that has been Reverse Transcriptase Polymerase Chain Reaction (RT-PCR), iso-thermal amplification, and CRISPR are 3 primary molecular approaches for identifying viral RNA. The most often utilised procedures for detecting coronavirus infection in samples of patient are PCR-based approaches, which includes traditional Reverse-Transcription PCR (RT-PCR) and Real-Time Quantitative PCR (qPCR) both [6].
WHO recommended Reverse Transcription Quantitative PCR (RT-qPCR) testing of samples of respiratory tract for laboratory confirmation and identification of cases of COVID-19 due to quality and safety check [7]. Many molecular targets within RNA Corona virus have come upon for use in PCR assay which includes helicase (Hel protein), nucleocapsid (N protein), trans-membrane (M protein), envelope (E protein) and glycoproteins spike (S protein). Other proteins in corona virus structure which are and can be used for the COVID-19 virus diagnosis includes
- Open reading frames which include ORF 1a and ORF 1b
- RNA-Dependent RNA polymerase (RdRp).
- Hemagglutinin-Esterase (HE)
In this rt-PCR assay, viral RNA is assessed using the Cycle Threshold (CT). Ct values for specimens are used to verify real-time PCR results; a value less than 40 is considered PCR positive in clinical practise. Although RT-PCR results are almost always 100 percent specific, false negative results might occur due to sample mistake or poor sampling timing [8]. As there is complete viral RNA extraction, reverse transcription, amplification, and availability of RT-PCR in hospital and institutes of research and labs, RT-PCR is regarded the best diagnostic choice for global surveillance. Other advantages of RT-PCR include the fact that it saves time, andare simple to use and is inexpensive. Depending on the type of RT-PCR methodology utilised, the total analytical procedure takes 4-8 hours (one-step or two-steps). This enables the result of a person tested positive in small interval of time, and hence fasten the process of quarantine and thus preventing disease in other individuals [9]. The WHO has issued specific suggestions beneficial for making an exact and proper diagnosis of viral RNA to decrease errors in analysis of molecular tests. In order to acquire valid results, Molecular test ought to be performed in 2 different biological substrates. For example swabs from 2 variable locations like a swab of nasopharynx and faecal swab, or on 2 successive swabs of nasopharynx from both the nostrils. Tests which are approved includes examination of viral genes three in number or viral genes two in number and control human gene, along with usage of controls positive and negative check presence of contamination or measure reaction inhibition [9]. One-step assays and two-step assays are the two most used methods for performing PCR test. One step test combines transcription (reverse) and PCR multiplication in single tube, makes detection procedure fast and of increased consistency. Although, the target amplicon generation is lower. Reaction is carried out in 2 steps in a sequence one undergoing after other in two different tubes. Therefore, it takes more time but its result is more sensitive when compared to a single step test [10]. Although RT-PCR can be used to provide a specific diagnosis of COVID-19, but it is less sensitive compared to chest CT scans but without a RTPCR test, a CT scan of chest cannot differentiate whether it is COVID-19 pneumonia or any other bacterial or viral pneumonia. There is another molecular test that is Loop Mediated Isothermal Amplification (LAMP) which is used for detecting COVID-19 as an option to RT-PCR and RT-qPCR. Loop-mediated isothermal amplification is an amplification method of nucleic acid and as name suggest multiplies DNA in a thermally stable (constant temperature) environment which provides greater specificity and speed. It can be a point-of-care device in future for COVID-19 diagnosis in infected people [8]. Rapid, simple, low-cost, portable, temperature-stable assays works on basis of Clustered Regularly Interspersed Short Palindromic Repeats that is CRISPR are also developed for deployment in the field in non-traditional and resource-limited situations, such as airports and border crossings (Figure 6) [11].
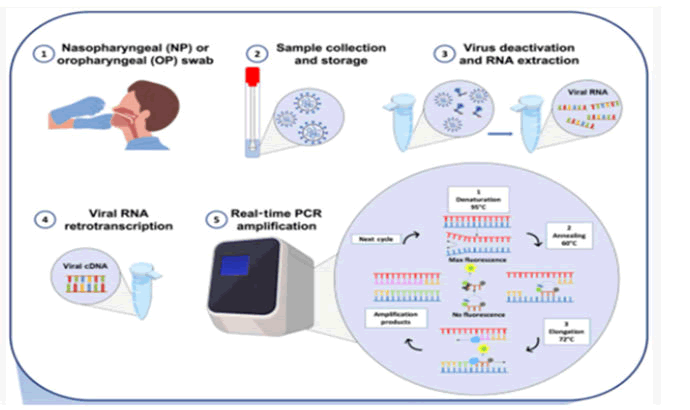
Figure 6: RT-PCR diagnosis workflow diagram. a) Nasopharyngeal or oropharyngeal swab collection; b) Swab handling and storage for viral RNA integrity; c)Viral RNA thermal inactivation and viral RNA extraction and d) retro-transcription of viral RNA e) Amplification.
Results and Discussion
Serological test
For detecting specific viruses, a variety of serological tests have been used, including Neutralisation tests,
- Immuno Fluorescent Assays (IFAs),
- Enzyme-Linked Immunosorbent Assays (ELISA),
- Immuno Chromatographic Tests (ICT).
Molecular testing is comparatively arduous than serological tests as serological test detects antibodies like IgA IgG and IgM antibodies from materials like blood and saliva by using assays like ELISA. Their diagnostic value for early infections must be restricted during symptoms onset, during this time, virus shedding appears to be at its peak, and the danger of transmission appears to be at its highest. For consistent antibody response many days and at times weeks are required. If results come out to be negative it doesn’t rule out COVID-19 infection, especially in those who have recently been exposed to the virus as it could be false negative. Antibody cross-reactivity with non-SARS-CoV-2 coronavirus proteins is also a concern, as positive results could be the consequence of previous or current infection with other human coronaviruses (Figures 7 and 8) [11].
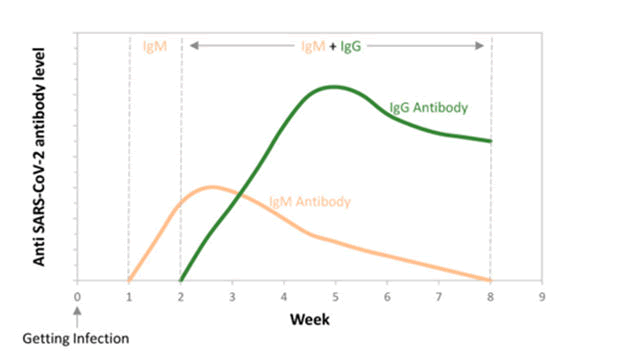
Figure 7: At different weeks following infection: initially there is no antibody formation. Then there is formation of IgM antibody and further IgG antibody formation occurs producing an immune response for longer time period.
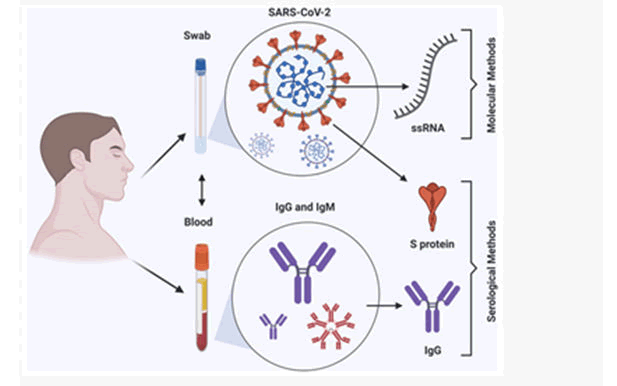
Figure 8: Summary: COVID-19 detection methods.
Enzyme-linked immunosorbent assay
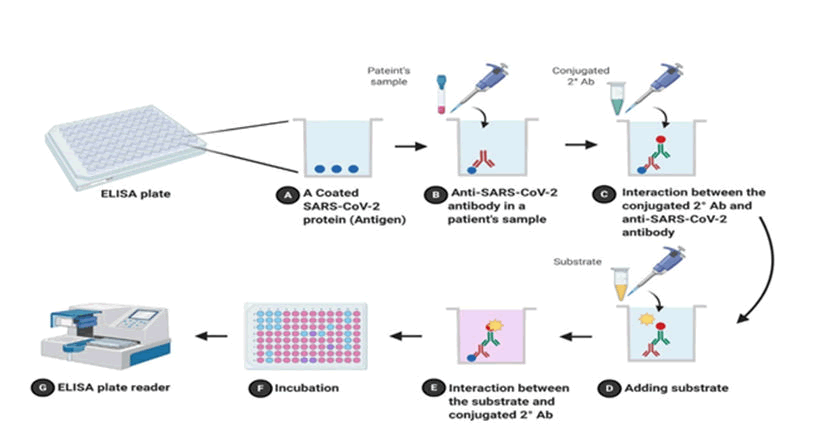
Enzyme Immunoassay is different type of serological test (EIA). In clinics and research laboratories, ELISA is for identifying and quantification of molecules which are soluble like proteins and antibodies. ELISA consists of direct test and indirect test both. Antigen is coated on inside surface of 96 well or 384 wells of polystyrene in indirect ELISA as it are more sensitive than direct ELISA. Wells are pre-filled with patient plasma which is diluted and it may consist of IgG/IgM antibody of SARS-CoV-2, after that plate is incubated and antibodies combine with antigens on surface for an hour. After washing the plate to remove unspecific contacts, a conjugated antibody that is antibody antigen complex with a recognised enzyme such as Horseradish Peroxidase (HRP) or Alkaline Phosphatase (AP) is added to make complexes. Then complexes are detected and estimated or quantification is done by introducing a substrate (for example: 3,3′,5,5′-tetramethylbenzidine) to the reaction. Results are shown as colour change in reaction. There is a plate reader for detecting and measuring colour. When compared to rRT-PCR, ELISA is quick (2-5 hours) and cheap. On day 0 of COVID-19 infection, ELISA results for patients were 49% (IgG) and 80% percent (IgM), and on day five, they were 80 percent (IgG) and 99 percent (IgM) (Figure 9 and Table 1).
Figure 9: Indirect ELISA flowchart diagram; a) Coated COVID antigen in well; b) Antibody in patients’ sample; c) Interaction between antigen and antibody; d) Addition of substrate e) Interaction of substrate and conjugated secondary antibody; f) Incubation; g) Reading of result on ELISA plate reader.
| Molecular methods | Serological methods | ||||
|---|---|---|---|---|---|
| Technique based | rRT-PCR | Isothermal amplification | CRISPR-Cas-12 | LFA | ELISA |
| Sample | RNA | RNA | RNA | Ag or Ab | Ag or Ab |
| Accuracy | High | High/Moderate | High | Low | Moderate |
| Time" | Hours | Minutes | Minutes | Minutes | Hours |
| Professional skills need | Yes | Yes | YES/No | No | Yes |
| POCT | No | Yes/No | Yes/No | Yes | No |
| Availability | Limited | Limited | Limited | Available | Available |
| Cost | Very high | High | Average | Low | Average |
| High throughput | Yes | No | No | No | Yes |
Table 1: Properties of NiTi and stainless steel rotary files.
Method: we have gathered knowledge from PubMed and google scholars using main term like diagnosis of COVID-19 considering both molecular and serological basis of virus detection. After gathering data from many articles this summary report is created. The global threat posed by the COVID-19 pandemic is serious issue. With a mild clinical manifestations and asymptomatic carriers, it's noticeable that controlling covid-19 requires an early and correct diagnosis. Rt-PCR is a critical component of SARS-CoV-2 laboratory diagnosis; however, it has significant drawbacks. A combine approach of Guiding patient management and infection control, combining laboratory tools (rt-PCR, serology and radiographic characteristics and clinical features) is required [12,13]. The present methods can be utilised quickly and mainly quantitatively in the event of a COVID-19 revival, allowing for the rapid detection of additional infected persons, isolation, and confinement measures. Further test optimization and clinical and epidemiological validation, as well as official FDA clearance, are still required [14,15]. Few of the related key studies were reviewed [16-21].
Conclusion
The diagnostic value of a test is not solely determined by the technique of collecting the material, the quality of the sample, or the equipment used, as it is with any other infectious disease. From the moment a sample is obtained, as well as a suitable technique (storage and handling) before to analysis, are equally important pre-analytical concerns. Author contribution: All authors made best contribution for the assessment and evaluation, concept, data acquisition and analysis and interpretation of the data.
References
- La Marca A, Capuzzo M, Paglia T, et al. Testing for SARS-CoV-2 (COVID-19): a systematic review and clinical guide to molecular and serological in-vitro diagnostic assays. Reprod Biomed Online 2020; 41:483-499.
- Kilic T, Weissleder R, Lee H. Molecular and immunological diagnostic tests of COVID-19: current status and challenges. Science 2020; 23:101406.
- Hassan SA, Sheikh FN, Jamal S, et al. Coronavirus (COVID-19): a review of clinical features, diagnosis, and treatment. Cureus 2020; 12.
- Beig Parikhani A, Bazaz M, Bamehr H, et al. The inclusive review on SARS-CoV-2 biology, epidemiology, diagnosis, and potential management options. Curr Microbial 2021; 78:1099-1114.
- Castagnoli R, Votto M, Licari A, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in children and adolescents: a systematic review. JAMA Pediatri 2020; 174:882-889.
- Mardian Y, Kosasih H, Karyana M, et al. Review of current COVID-19 diagnostics and opportunities for further development. Front Med 2021; 8:562.
[Crossref][Google Scholar][Pubmed]
- Zheng Z, Yao Z, Wu K, et al. The diagnosis of SARSâ?CoV-2 pneumonia: A review of laboratory and radiological testing results. J Med Virol 2020; 92:2420-2428.
- Kubina R, Dziedzic A. Molecular and serological tests for COVID-19. A comparative review of SARS-CoV-2 coronavirus laboratory and point-of-care diagnostics. Diagnostics 2020; 10:434.
- Mathuria JP, Yadav R. Laboratory diagnosis of SARS-CoV-2-A review of current methods. J Infect Public Heal 2020; 13:901-915.
- Falzone L, Gattuso G, Tsatsakis A, et al. Current and innovative methods for the diagnosis of COVIDâ??19 infection. Int J Mole Med 2021; 47:1-23.
- Islam KU, Iqbal J. An update on molecular diagnostics for COVID-19. Front Cell Infect Microbiol 2020; 694.
- Cheng MP, Papenburg J, Desjardins M, et al. Diagnostic testing for severe acute respiratory syndromeâ??related coronavirus 2: a narrative review. Ann Int Med 2020; 172:726-734.
- Taleghani N, Taghipour F. Diagnosis of COVID-19 for controlling the pandemic: A review of the state-of-the-art. Biosens Bioelectron 2021; 174:112830.
- Dhamad AE, Rhida MA. COVID-19: molecular and serological detection methods. Peer J 2020; 8:e10180.
- Vandenberg O, Martiny D, Rochas O, et al. Considerations for diagnostic COVID-19 tests. Nat Rev Microbiol 2021; 19:171-183.
[Crossref][Google Scholar][Pubmed].
- Godhiwala P, Acharya S, Jagtap G, et al. Leukemoid Reaction in a COVID-19 Patient. J Evo Med Dent Sci 2021; 8:399-400.
- Taksande A. Myocardial dysfunction in SARS-CoV-2 infection in infants under 1 year of age. World J Pediatrics 2020; 16:539.
- Jaiswal G, Jha RK, et al. Indian Laws during COVID-19 Pandemic: Implications to Health and Management. J Pharma Res Int 2021; 232-236
- Madaan S, Jaiswal A, Kumar S, et al. Cytokine storm treated successfully with immunoglobulin therapy in a pregnant COVID-19 patient. Med Sci 2021; 25:1413-1416
- Hardaswani D. COVID-19, a Pandemic Situation: Symptoms and its Prevention Scenario. J Pharma Res Int 2021; 33.
- Bidkar V, Selvaraj K, Mishra M, et al. A comparison of swab types on sample adequacy, suspects comfort and provider preference in COVID-19. Ame J Otolaryngo 2021; 42:102872.
Author Info
Pranjali Jain* and Swarupa Chakole
Department of Community Medicine Jawaharlal Nehru Medical College, Datta Meghe Institute of Medical Sciences (DU), Sawangi Meghe, Wardha, Maharashtra, IndiaCitation: Pranjali Jain, Swarupa Chakole Testing for SARS-COV-2: Molecular and Serological diagnosis J Res Med Dent Sci, 2022, 10 (7): 000-000
Received: 27-May-2022, Manuscript No. JRMDS-22-49621; , Pre QC No. JRMDS-22-49621; Editor assigned: 31-May-2022, Pre QC No. JRMDS-22-49621; Reviewed: 14-Jun-2022, QC No. JRMDS-22-49621; Revised: 18-Jul-2022, Manuscript No. JRMDS-22-49621; Published: 06-Aug-2022