Research - (2020) Volume 8, Issue 4
Recombination Analysis of S-Segment Genome of Crimean-Congo Hemorrhagic Fever Virus in Turkey
Murtaza Oklu1, Ibrahim Suslu2, Cengiz Z Altuntas3 and Senol Dane4*
*Correspondence: Senol Dane, Department of Physiology, Faculty of Basic Medical Sciences, College of Health Sciences, Nile University of Nigeria, Abuja, Nigeria, Email:
Abstract
Background: Crimean-Congo Hemorrhagic Fever (CCHF) is an illness that caused by a tick-borne virus, which belongs to family Bunyaviridae, genus Nairovirus. It is carried on migratory birds. CCHF disease has been appeared in Europe, Asia, and Africa. Then, the disease spread in other cities in Turkey such as Yozgat, Eskisehir, Kelkit, and Corum. RNA recombination is mixing of genetic material strains from different countries into new combinations that cause RNA virus diversity. The aim of this study is the recombination analysis of complete S-segment genome among CCHFV strains in Turkey and strains from different countries.
Materials and Methods: S-segment has the strongest geographical subdivision, and the investigation S-segment genome of CCHFV explained that there are differences in nucleotide sequences between strains. Turkey is a center for distribution of migratory birds. In this study, we compared strains whit complete sequence from Russia, Bulgaria, Kosovo, Greece, Iran, Nigeria, Mauritania, Sudan, South Africa, and Mali, where have migratory birds’ flyways. The GenBank information is downloaded from www.ncbi.nlm.nih. gov in FASTA format and investigated with phylogenetic tree. Evidence of recombination is analyzed by using SimPlot and MEGA X software.
Results: The strains in Corum, Kelkit, Eskisehir, Yozgat are mainly recombinant of the strains in Iran, Russia, Bulgaria, and Nigeria in orderly.
Conclusion: This research revealed that African-Eurasian migratory bird flyways provide the way for intercontinental migration of the CCHFV that causes recombination of CCHFV in Turkey. Thus, the vaccine design should be updated as concerned of recombination of CCHFV.
Keywords
Recombination, Crimean Congo Hemorrhagic Fever, Turkey, S-segment, Phylogenetic
Introduction
CCHF is an illness that caused by a tick-borne virus, which belongs to family Bunyaviridae, genus Nairovirus. The virus is carried on migratory birds [1,2]. CCHF disease has been appeared in Europe, Asia, and Africa and distributed from Africa, Asia, and Europe to wide range [3]. In 2002, CCHF virus cases were reported at the first time in Tokat, Turkey [4]. Then, the disease was detected from other cities such as Corum, Aydin, Yozgat, Kayseri, Eskisehir, Kayseri, and Gumushane. Only humans and sucking mice has affected from the disease and there are no clinical symptoms in other vertebrates [5]. The disease is transferred to human body with bites from infected ticks. CCHF is an important health issue around the world.
The infected ticks are carried on migratory birds and spread to enormous area on their migration flyways. The spreading of CCHF virus is caused genetic varieties which can be explained by recombination, reassortment, and mutation [3,6]. RNA viruses have ability to change their population size and high rates of mutation quickly [7]. Many data are collected from different cities and investigated to learn recombination between strains in Turkey, and the results showed that there is genetic diversity between infected ticks. Also, in our study we compared the genomes of CCHF viruses from different countries, which are chosen about of recombination relation with Turkey, such as Russia, Bulgaria, Kosovo, Greece, Iran, Nigeria, Mauritania, Sudan, South Africa, and Mali. The reason of analyzing CCHF virus from different countries is to understand better how infected virus spread in an enormous area. The aim of this study is the recombination analysis of complete S-segment genome among CCHFV strains in Turkey and strains from different countries.
Materials and Methods
Recombination has a significant impact for designing genetic differences in RNA viruses [8]. CCHF virus contains three segments, which are S, M, and L, genome and depend on phylogenetic analysis, S-segment has the strongest geographical subdivision [9]. The result of investigated the S-segment genome of CCHFV explained that there are differences in nucleotide sequences between cities. The GenBank information is downloaded from www.ncbi.nlm. nih.gov in FASTA format and investigated with phylogenetic tree. Evidence of recombination is analyzed by using SimPlot and MEGA X software. SimPlot program performed boot scan method for analyzing the recombination of the strains and shows the consequences with a graphical figure. MEGA X software program is used the determine the situation of the strains in a phylogenetic tree. First, the strains are chosen from the cities of Turkey, then the nucleotide-blast method is used to determine which strains have similarities with the chosen strains. Then, the data of the strains are collected in a file for using in SimPlot and MEGA X software. The data is used in MEGA X software to find out phylogenetic tree of the chosen strains. After that, the data is transferred to SimPlot program to explore which strains have recombination with each other. The strains are marked with different colors to determine and understand the graphic.
Results
The strains are supplied from www.ncbi.nlm. nih.gov for detecting the recombination between each other. Crimean-Congo hemorrhagic fever virus is spread around the world from different countries with immigrant birds. A strain has been found from a patient in Turkey was genetically quite different than the previous strains which found in Tukey, but the strain is related with a virus which found in Greece [10]. For this project, some strains are collected from various countries such as Turkey, Russia, Bulgaria, Nigeria, Mauritania, Sudan, Iran, Greece, and Kosovo. Name of the viruses, countries, year of isolations, and GenBank accession numbers are given in Table 1 to show the data of the strain in Table 1. The strains in Corum, Kelkit, Eskisehir, Yozgat, which Turkish cities, are mainly recombinant of the strains in Iran, Russia, Bulgaria, and Nigeria in orderly.
| Name of virus isolate | Country | Year of isolation | GenBank accession no S-segment |
|---|---|---|---|
| Turkey-Kelkit 06 | Turkey | 2006 | GQ337053 |
| Yozgat 19-2012 | Turkey | 2012 | KR092375 |
| Eskisehir 23-2012 | Turkey | 2012 | KR092376 |
| Corum 1094-2011 | Turkey | 2011 | KR092378 |
| 9553/2001 | Kosovo | 2001 | AF428144 |
| Kosovo Hoti | Kosovo | 1980 | DQ133507 |
| V42/81 | Bulgaria | 1981 | GU477489 |
| ROS/TI28044 | Bulgaria | 2003 | AY277672 |
| 66/08-Rodopi | Greece | 2008 | EU871766 |
| 46-ST-2012 | Russia | 2012 | KR814841 |
| Kashmanov | Russia | 1967 | DQ211644 |
| ROS/HUVLV-100 | Russia | 2002 | DQ206447 |
| Ast133 | Russia | 2012 | KX056061 |
| Iran-Isfahan78 | Iran | 2013 | KJ027522 |
| Iran-Gilan69 | Iran | 2012 | KJ027521 |
| Iran-Kerman43 | Iran | 2013 | KJ196326 |
| Iran-Tehran65 | Iran | 2011 | KJ566219 |
| Iran-Zahedan19 | Iran | 2012 | KJ676542 |
| UCCR4401 | Nigeria | 1996 | KY484036 |
| ArD39554 | Mauritania | 1984 | DQ211641 |
| Sudan_Al_Fulah_3-2008 | Sudan | 2008 | GQ862371 |
Table 1: Crimean-Congo Hemorrhagic Fever virus isolates used in this study, with related countries of origin, collection date, and S-segment GenBank accession numbers.
The result of Figure 1A, from SimPlot shows that the strains from Yozgat, Nigeria, Bulgaria, and Russia have recombination between each other. Yozgat strain is used as query for finding out recombination possibilities with the others. TURKEY-YOZGAT19-2012-KR092375 has recombination with, NIGERIA-UCCR4401- KY484036 in position of 450-630 and 1,150- 1,370, BULGARIA-V42/81-GU477489 in position of 150-200, 250-430, 730-850, and 930-1,150, and RUSSIA-AST133-KX056061 in position of 1,370-1,570.
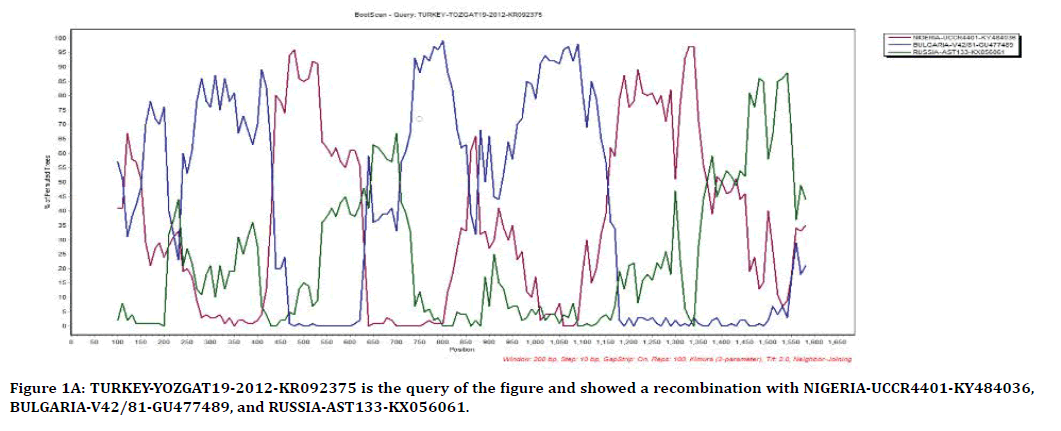
Figure 1A. TURKEY-YOZGAT19-2012-KR092375 is the query of the figure and showed a recombination with NIGERIA-UCCR4401-KY484036, BULGARIA-V42/81-GU477489, and RUSSIA-AST133-KX056061.
The result of Figure 1B from SimPlot shows that the strains from Corum, Mauritania, Bulgaria, and Russia have recombination between each other. Corum strain is used as query for finding out recombination possibilities with the others. TURKEY-CORUM1094_2011-KR092378 has recombination with, MAURITANIA-ARD39554- DQ211641 in position of 420-680 and 1,100- 1,450, BULGARIA-AY277676 in position of 100- 220 and 700-900, RUSSIA-ROS/HUVLV-100- DQ206447 in position of 220-420, 900-1,100, and 1,450-1,580.
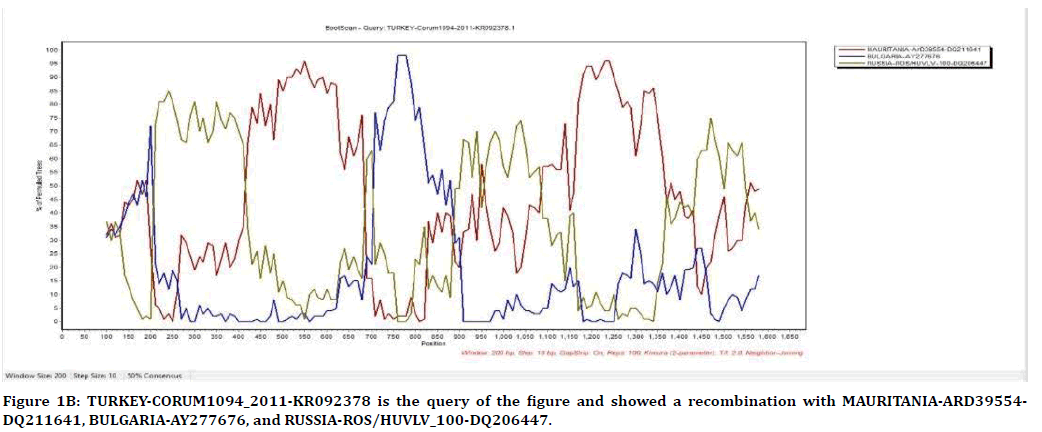
Figure 1B. TURKEY-CORUM1094_2011-KR092378 is the query of the figure and showed a recombination with MAURITANIA-ARD39554-DQ211641, BULGARIA-AY277676, and RUSSIA-ROS/HUVLV_100-DQ206447.
The result of Figure 1C from SimPlot shows that the strains from Eskisehir, Russia, Iran, and Greece have recombination between each other. Eskisehir strain is used as query for finding out recombination possibilities with the others. TURKEY-ESKISEHIR23-2012-KR092376 has recombination with RUSSIA-AST133-KX056061 in position of 200-420, 470-500, 550-820, and 1,450-1,550, IRAN-ZAHEDAN19-KJ676542 in position of 820-1,100, and GREECE-66/08RODOPIEU871766 in position of 1,100-1,450.
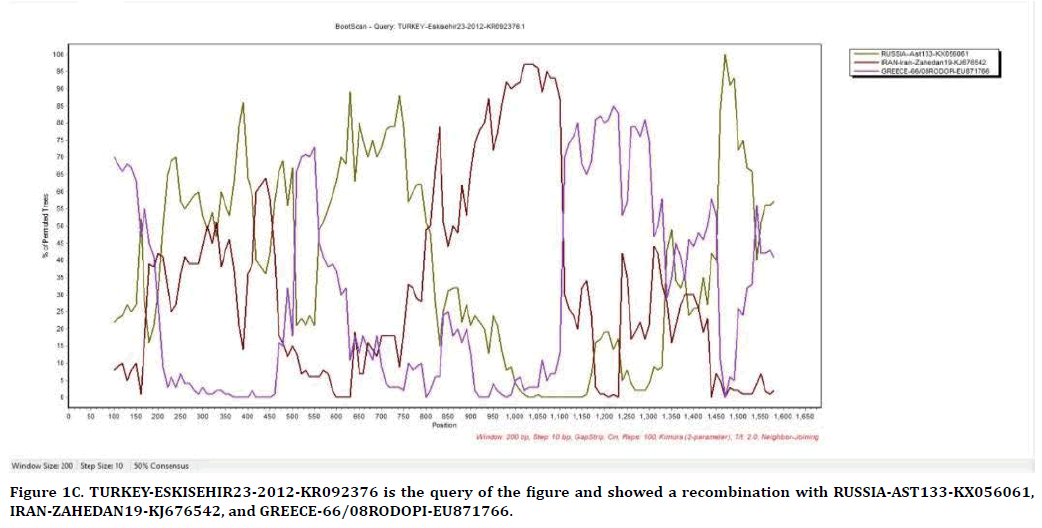
Figure 1C. TURKEY-ESKISEHIR23-2012-KR092376 is the query of the figure and showed a recombination with RUSSIA-AST133-KX056061, IRAN-ZAHEDAN19-KJ676542, and GREECE-66/08RODOPI-EU871766.
The result of Figure 1D from SimPlot shows that the strains from Kelkit, Russia, Sudan, and Kosovo have recombination between each other. Kelkit strain is used as query for finding out recombination possibilities with the others. TURKEY-KELKIT06-GQ337053 has recombination with KOSOVO-9553/2001- AF428144 in position of 200-250 and 780-1,180, SUDAN-SUDAN_AL-FULAH_3-2008-GQ862371 in position of 440-560, 1,180-1,250, and 1,320- 1,520, and RUSSIA-KASHMANOV-DQ211644 in position of 100-200, 250-450, 570-770, 950- 1,050, and 1,500-1600.
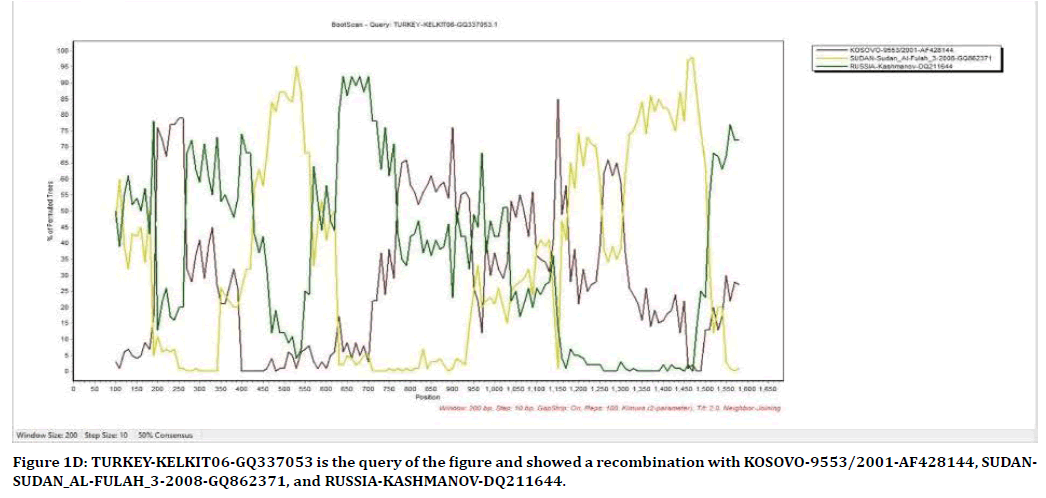
Figure 1D. TURKEY-KELKIT06-GQ337053 is the query of the figure and showed a recombination with KOSOVO-9553/2001-AF428144, SUDANSUDAN_ AL-FULAH_3-2008-GQ862371, and RUSSIA-KASHMANOV-DQ211644.
In Figure 1e, phylogenetic tree shows that how the strains from various countries close each other by using MEGA X Software. Strains from the same countries have more similarities than others because of that they have close relation with each other.

Figure 1E. The strains from different countries compared in a phylogenetic tree by using MEGA X software.
Discussion
Nowadays, a big increase is seen in the incidence of CCHF disease in Turkey. The results of the present study support that the tick viruses are migrated from northwestern Africa to Europe by migratory birds [11]. The migratory birds’ flyways have a significant effect to spreading of CCHF virus. The analyzes of strains from different cities in Turkey have similarities on strains from other countries because of recombination from infected viruses that carried on migration bird flyways.
This research revealed that African-Eurasian migratory bird flyways provide the way for intercontinental migration of the CCHFV that might cause recombination of CCHFV in Turkey. Considering the recombination and reassortment in CCHF virus indicates that common isolates in the area has been altered in genetical and serological way from the viruses [5]. Thus, the vaccine design should be updated as concerned of recombination of CCHFV.
Conclusion
Analysis of complete S-segment genome among CCHFV strains in Turkey and strains from different countries revealed that all strains in Turkey have recombination.
References
- Watts DM, Ksiazek TG, Linthicum KJ, et al. Crimean-congo hemorrhagic fever. In: Monath TP. The arboviruses epidemiology and ecology. Boca Raton, USA: CRC Press 1988; 2:177-260.
- Leblebicioglu H, Eroglu C, Erciyas-Yavuz K, et al. Role of migratory birds in spreading Crimean-Congo hemorrhagic fever, Turkey. Emerg Infect Dis 2014; 20:1331-1334.
- Chinikar S, Shah-Hosseini N, Bouzari S, et al. Assessment of recombination in the S-segment genome of crimean-congo hemorrhagic fever virus in Iran. J Arthropod Borne Dis 2015; 10:12-23.
- Karti SS, Odabasi Z, Korten V, et al. Crimean congo hemorrhagic fever in Turkey. Emerg Infect Dis 2004; 10:1379-1384.
- Lukashev AN. Evidence for recombination in crimean congo hemorrhagic fever virus. J Gen Virol 2005; 86:2333-2338.
- Deyde VM, Khristova ML, Rollin PE, et al. Crimean congo hemorrhagic fever virus genomics and global diversity. J Virol 2006; 80:8834-8842.
- Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annu Rev Microbiol 1997; 51:151-178.
- Chare ER, Gould EA, Holmes EC. Phylogenetic analysis reveals a low rate of homologous recombination in negative-sense RNA viruses. J Gen Virol 2003; 84:2691-2703.
- Zhou Z, Deng F, Han N, et al. Reassortment and migration analysis of Crimean congo haemorrhagic fever virus. J Gen Virol 2013; 94: 2536-2548.
- Elevli M, Ozkul AA, Civilibal M, et al. A newly identified crimean-congo hemorrhagic fever virus strain in Turkey. Int J Infect Dis 2010; 14:213-216.
- Palomar AM, Portillo A, Santibáñez P, et al. Crimean congo hemorrhagic fever virus in ticks from migratory birds, Morocco. Emerg Infect Dis 2013; 19:260-263.
Author Info
Murtaza Oklu1, Ibrahim Suslu2, Cengiz Z Altuntas3 and Senol Dane4*
1Department of Computer Science, University of Houston-Downtown, Houston, TX 77002, USA2Department of Computer Science, North American University, Houston, TX 77477, USA
3Texas Institute of Biotechnology Education and Research (TIBER), North American University, Houston, TX 77477, USA
4Department of Physiology, Faculty of Basic Medical Sciences, College of Health Sciences, Nile University of Nigeria, Abuja, Nigeria
Citation: Murtaza Oklu, Ibrahim Suslu, Cengiz Z Altuntas, Senol Dane, Recombination Analysis of S-Segment Genome of Crimean-Congo Hemorrhagic Fever Virus in Turkey, J Res Med Dent Sci, 2020, 8 (4):141-145x.
Received: 26-Jun-2020 Accepted: 20-Jul-2020
