Research - (2021) Volume 9, Issue 11
Identification of Potential Metabolites in Chlorella Species and Oscillatoria Species for Treatment of Multidrug Resistant (MDR) Infections: An In Silico Analysis
*Correspondence: Abubucker Peer Mohideen, Department of Basic Medical Sciences, College of Medicine, Prince Sattam Bin Abdulaziz University, Al-Kharj, 11942, Saudi Arabia, Email:
Abstract
The emergence of new infectious agents with antimicrobial resistance (AMR) is threatening the whole world. Multidrug resistant (MDR) bacterial infections are found to be an alarming global crisis with limited availability of antibiotics. The MDR co-infection among COVID-19 infected patients led to higher mortality rates, which increased the necessity for development of novel antibacterial agents with a broad spectrum of activity. Microalgae metabolites serve as an untapped potential source with a wide range of biological activities. The present study concentrates on the identification of novel antibacterial microalgal metabolites in Oscillatoria sp and Chlorella sp. The compounds were retrieved from the KNApSAck database. Tyrosyl-tRNA synthetase and DNA Gyrase were used as the dual target proteins. Molecular docking and protein-ligand interactions were performed to identify the compounds with higher binding energies (>-7 kcal/mol). Further, the compounds were analyzed for Druglikness (RO5), PASS Predictions and ADMET analysis. From the analysis, 12, 13-Trans-Epoxy-9-oxo-10E, 15Z-octadecadienoic acid, Ergosta-8, (9) 14-dien-3beta-ol, Jasmonic acid, Methyl jasmonate, and Poriferasterol were identified with potential binding energy, interactions, and drug likeness properties. All the selected compounds showed significant ADME properties when compared with the standard drug Azithromycin and were found to be non-toxic/carcinogen. In conclusion, the identified six compounds can be utilized for the development of novel antibacterial drugs for the treatment of MDR infections.
Keywords
Multidrug resistant (MDR) bacterial infections, COVID 19 co-infections, Molecular docking, PASS Predictions, ADMET analysis
Introduction
The worldwide public health is greatly threatened by antibiotic-resistant infections, particularly those produced by multi-drug-resistant (MDRO) organisms. The indiscriminate use of antibiotics has led to a growing pharmacology resistance amongst bacterial infections, and a global crisis has been proclaimed by the World Health Organization (WHO).
In patients with past antibiotic exposure or comorbidity, critically sick patients are especially susceptible to MDRO infection, which can increase mortality, hospitalisation and hospital stays. Despite the negative impact of antibiotic resistance on patients worldwide, resistance burden analyses have been conducted in low-and middle-income (LMIC) countries, even through increased income, lower pharmaceutical costs, and unregulated sales, leading to increased antibiotic use and higher rates of resistance [1].
The US results in more than 35,000 fatalities each year due to antibiotic resistance and more than 2 month hospital stays. In the EU, 25,000 fatalities and an additional 2.5 months in hospital are caused by antibiotic resistance. More than 58,000 infants perished in one year from infections in India, generally transmitted by their mothers, with resistant bacteria. Thailand has 38,000+ fatalities and 3.2 million hospital days a year due to antibiotic resistance.
With their particular mechanisms of resistance leading to ineffective medicines, several drug-resistant bacterial diseases have emerged. The principal methods of drug resistance are inactivation of or altering medication by many means, such as enzyme interventions, change of drug sites, reduced intracellular accumulation of drugs, development of biofilms, etc. New antimicrobial medicines are therefore needed to treat such bacterial MDR infections [2,3].
Microalgae are minuscule, freshwater, estuarine, and marine organisms that may be found in many habitats. Usually, they are single-cell micro-organisms and can be in chains or groups alone. Their size can vary from a few micrometres to a few hundred micrometres, depending on species. Microalgae serve as a valuable source for many bioactive compounds responsible for various biological activities [4].
They must evolve tolerance/defenses mechanisms to live against other pathogenic microorganisms. These techniques have led to the synthesis of antibacterial compounds with a significant variety and diverse mechanisms of action. Since they have a very basic structure and a high surface/volume ratio, which results in a high nutrient assimilation rate, the growth of green algae is about one hundred times as quick as the growth of terrestrial plants. The rapid growth and simple nutrient requirements attracted researchers to explore the potential of microalgae species all over the world [5].
Therefore, the present study is a screening process of identification of microalgal metabolites having antibacterial properties through in silico analysis. The compounds from Oscillatoria sp. and Chlorella sp. were retrieved and docked against dual bacterial targets DNA Gyrase and DNA topoisomerase. Further the compounds were subjected to drug likeness, PASS predictions and ADMET analysis.
Materials and Methods
Target proteins preparation
The tyrosyl-tRNA synthetase (PDB ID: 1JIJ) and DNA Gyrase subunit b (PDB ID: 1KZN) are utilized as target proteins in this study and their 3D structures were acquired from the Protein Data Bank (http:// www.rcsb.org/). The target proteins are visualized using PyMol tool and the protein associated water molecules, ligands and co-crystal ligands were removed (Figure 1).
Figure 1: 3D structures of the Target proteins. (a) Tyrosylt RNA synthetase (b) DNA Gyrase.
The proteins were prepared in Auto Dock Tools, open source free software by adding charges and energy minimization was carried out in Swiss PDB viewer and further converted to pdbqt format.
Retrieval and preparation of ligands
The bioactive compounds present in Oscillatoria sp. and Chlorella sp. were identified and retrieved using KNApSAck database (http://www.knapsackfamily.com/ KNApSAcK/). Total of 39 bioactive compounds were used for the study (Table 1). The preparation of ligand is performed by identifying its torsion root, assigning charges, correcting torsion angle, optimising using UFF (universal force field) and lastly converting into pdbqt format to create 3D atomic coordinates of the molecules [6].
| S. No | Compound name | S. No | Compound name |
|---|---|---|---|
| 1 | beta-Echinenone | 21 | Largamide B |
| 2 | Anatoxin alpha | 22 | Largamide C |
| 3 | Mutatochrome | 23 | Largamide D |
| 4 | Aphanizophyll | 24 | Largamide E |
| 5 | 3'-Hydroxyechinenone | 25 | Largamide F |
| 6 | Aeruginosin 205A | 26 | Largamide G |
| 7 | Aeruginosin 205B | 27 | Largamide H |
| 8 | Anabaenopeptin F | 28 | Viridamide A |
| 9 | Anabaenopeptin G | 29 | Homoanatoxin-a |
| 10 | Anabaenopeptin H | 30 | 12,13-trans-Epoxy-9-oxo-10E,15Z-octadecadienoic acid |
| 11 | Lipopurealin A | 31 | 13-Hydroperoxyoctadecadienoic acid |
| 12 | Oscillamide H | 32 | 13-Hydroxyoctadecadienoic acid |
| 13 | Oscillamide Y | 33 | 9-Hydroperoxyoctadecadienoic acid |
| 14 | Prenylagaramide A | 34 | Ergosta-8,(9)14-dien-3beta-ol |
| 15 | Prenylagaramide B | 35 | Jasmonic acid |
| 16 | Raocyclamide A | 36 | Linolenic acid |
| 17 | Anabaenopeptin B | 37 | Loroxanthin |
| 18 | Microviridin I | 38 | Methyl jasmonate |
| 19 | Oscillacyclin | 39 | Poriferasterol |
| 20 | Largamide A |
Table 1: Compounds used in the study.
Determination of functional sites of targets
Significant docking analysis requires precise evaluation of the active (Functional) site. The amino acid residues in the active pocket site formation of target proteins was identified using the CAST ponline server (Computed Atlas for Surface Topography) [7,8].
CASTp is a simple and handy tool to analysing protein topology and active site pockets. Determining the active site is a critical for setting the grid box prior to docking.
Molecular docking and protein-ligand interaction analysis
All the compounds were docked using the PyRx tool via auto dock wizard. Throughout the docking process, it was believed that the ligands were flexible and the protein was rigid.
The grid parameter configuration file is created in PyRx using the grid boxes for 6W41 (x=-12.59, y=-18.04, z=83.05) and 6LU7 (x=17.27, y=30.68, z=48.04) [9].
Followed by docking, the ligand with the greatest binding energy (most negative) was identified as having the highest binding affinity.
The ligands with a greater binding energy (-7 Kcal/mol) were identified, and the interaction between the ligand and the protein at the binding sites was analysed using Biovia Drug discovery studio 2019.
Screening of the of ligands for drug likeness
Swiss ADME (http://swissadme.ch/index.php) is used to evaluate the compound’s drug likeness. The drug likeness of a molecule is a critical criterion for validating it as a possible agonist for therapeutic targets.
Compounds showing higher binding energy (<-7 Kcal/ mol) were screened for Druglikness property using the Lipinski’s Rule of Five (RO5) [10]. Prediction of activity spectra for substances (PASS) for antibacterial activity
The antibacterial activity of the identified compounds was predicted using the Prediction of activity spectra for substances (PASS) programme [11]. PASS programme is used to predict a variety of physiological effects for a large number of substances.
The substance's activity is predicted and quantified as probable activity (Pa) and probable inactivity (Pi). The only components that have a Pa greater than Pi are those that are viable for the particular medical activity.
ADMET analysis
ADMET analysis utilises online-based algorithms to determine the Absorption, Distribution, Metabolism, Excretion, and Toxicity of particular compounds.
Numerous online databases and offline software programmes are available to aid in the prediction of drug candidate behaviour.
We used admit SAR to make ADMET predictions in this investigation [12]. The compounds with the highest binding energies were evaluated for their intestinal absorption, blood-brain barrier penetration, in vitro Caco-2 cell permeability, CYP450 2C9 substrate, and toxicity characteristics such as mutagenicity as assessed by the Ames test and carcinogenicity in rats.
Ciprofloxacin, a broad spectrum antibiotics used as a reference drug on which the compounds are compared.
Results and Discussion
Binding site analysis and molecular docking
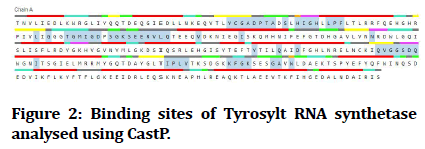
CASTp was used to determine the functional site pockets in tyrosyl-tRNA synthetase and DNA Gyrase. CASTp is a web-based tool for determining the amino acid residues in a protein's active pocket.
Figure 2 illustrates the CASTp results for tyrosyl-tRNA synthetase and Figure 3 illustrates the CASTp results for DNA Gyrase. From CASTp results, only the amino acids in the active site and their positions are listed as Table 2.
Figure 2: Binding sites of Tyrosylt RNA synthetase analysed using CastP.
| S. No | Target Protein | Amino acid residues in binding sites |
|---|---|---|
| 1 | Tyrosyl-tRNA synthetase (PDB ID: 1JIJ) | TYR-36, CYS-37, GLY-38, ALA-39, ASP-40, PRO-41, THR-42, ALA-43, SER-45, HIS-47, ILE-48, GLY-49, HIS-50, LEU-52, PRO-53, PHE-54, LEU-70, GLY-72, THR-75, GLY-76, MET-77, ILE-78, GLY-79, ASP-80, SER-82, GLY-83, LYS-84, SER-85, GLU-86, GLU-87, ARG-88, VAL-89, LEU-90, GLN-91, VAL-96, ILE-103, ASN-124, TYR-170, GLN-174, ASP-177, GLN-190, 191-VAL, GLY-192, GLY-193, SER-194, ASP-195, GLN-196, ILE-200, ILE-221, PRO-222, LEU-223, VAL-224, 231-LYS, PHE-232, GLY-233, LYS-234, GLY-238, ALA-239, TRP-241 |
| 2 | DNA gyrase (PDB ID: 1KZN) | GLU-58, ILE-60, GLN-72, ASP-73, ASP-74, VAL-133, GLN-135, LYS-162, THR-163, GLY-164, THR-165, MET-166 |
Table 2: Amino acid residues in the active sites.
Figure 3: Binding sites of DNA Gyrase analysed using CastP.
Grid box were generated covering the binding sites of the target protein.
PyRx was utilised to dock all 39 compounds to their target proteins, tyrosyl-tRNA synthetase and DNA Gyrase. The binding energies of the compounds were determined, and those with a lower binding energy (-7.0 Kcal/mol) against dual targets were identified. Around 19 compounds demonstrated a substantial binding energy (-7.0 Kcal/mol) for both targets and compounds, which are listed in Table 3.
| S. No | Microalgae compounds | Binding energy (kcal/mol) | |
| 1JIJ | 1KZN | ||
| 1 | beta-Echinenone | -6.4 | -7.1 |
| 2 | Anatoxin alpha | -5.8 | -5.5 |
| 3 | Mutatochrome | -6.9 | -7.5 |
| 4 | Aphanizophyll | 21.1 | -6.5 |
| 5 | 3'-Hydroxyechinenone | -6.2 | -7.3 |
| 6 | Aeruginosin 205A | -8.6 | -6.4 |
| 7 | Aeruginosin 205B* | -8.7 | -7.6 |
| 8 | Anabaenopeptin F* | -8.6 | -8 |
| 9 | Anabaenopeptin G* | -9.3 | -7.3 |
| 10 | Anabaenopeptin H* | -9.5 | -7.4 |
| 11 | Lipopurealin A | -7.6 | -6.2 |
| 12 | Oscillamide H* | -8.5 | -8.3 |
| 13 | Oscillamide Y* | -8.6 | -9.6 |
| 14 | Prenylagaramide A | -6.3 | -8.1 |
| 15 | Prenylagaramide B* | -9 | -8.2 |
| 16 | Raocyclamide A* | -9.1 | -7.6 |
| 17 | Anabaenopeptin B* | -8.1 | -9 |
| 18 | Microviridin I | 9 | -7.5 |
| 19 | Oscillacyclin | -5.7 | -7.6 |
| 20 | Largamide A* | -9.8 | -8.6 |
| 21 | Largamide B* | -9.3 | -7.8 |
| 22 | Largamide C* | -9.4 | -8.3 |
| 23 | Largamide D* | -6.2 | -6.8 |
| 24 | Largamide E* | -7.2 | -6.4 |
| 25 | Largamide F* | -6.2 | -7.7 |
| 26 | Largamide G* | -9.6 | -8.4 |
| 27 | Largamide H | -1.4 | -6.5 |
| 28 | Viridamide A | -7.2 | -5.4 |
| 29 | Homoanatoxin-a | -6 | -5.9 |
| 30 | 12, 13-trans-Epoxy-9-oxo-10E, 15Z-octadecadienoic acid* | -7 | -7.3 |
| 31 | 13-Hydroperoxyoctadecadienoic acid* | -8.3 | -7.7 |
| 32 | 13-Hydroxyoctadecadienoic acid | -6.3 | -5.4 |
| 33 | 9-Hydroperoxyoctadecadienoic acid | -6.5 | -5.7 |
| 34 | Ergosta-8,(9)14-dien-3beta-ol* | -7.8 | -8.3 |
| 35 | Jasmonic acid* | -7.7 | -7 |
| 36 | Linolenic acid | -5.8 | -5.1 |
| 37 | Loroxanthin* | -6.4 | -7.5 |
| 38 | Methyl jasmonate* | -7.5 | -7.8 |
| 39 | Poriferasterol* | -8.5 | -7.8 |
| *Compounds showing binding energies higher than -7 Kcal/mol | |||
Table 3: Binding energies of the compounds against the target proteins.
Analysis of protein-ligand interaction
The best-docked compounds were further examined for binding interactions on active sites with amino acid residues using Biovia Accelrys Discovery Studio Visualizer software. Bonding type, number of hydrogen bonds and hydrophobic interactions are a very important determinant of protein-ligand interactions as equal as binding affinity. The number of hydrogen bonds formed and amino acids involved in the interactions are tabulated in Table 4. The hydrogen bonds and other hydrophobic interactions of the ligands on the binding sites of the target proteins were shown in the Figure 4. All the 19 compounds showed H-bond formation on binding sites of the target proteins except Oscillamide Y, Prenylagaramide B, Largamide B, Largamide C and Largamide G. These compounds failed to show H-bond on binding sites of DNA Gyrase.
| S. No | Microalgae compound | 1JIJ | 1KZN | ||
|---|---|---|---|---|---|
| No of H-bond | Binding amino acid residue | No of H-bond | Binding amino acid residue | ||
| 1 | Aeruginosin 205B | 8 | A: 47-HIS; 50-HIS; 84-LYS; 193-GLY; 195-ASP; 233-GLY | 1 | A: 165-THR |
| 2 | Anabaenopeptin F | 5 | A: 47-HIS; 50-HIS; 89-VAL; 91-GLN | 5 | A: 72-GLN; 73-ASP; 135-GLN; 165-THR |
| 3 | Anabaenopeptin G | 4 | A: 47-HIS; 50-HIS; 88-ARG; 174-GLN | 2 | A: 165-THR; 73-ASP |
| 4 | Anabaenopeptin H | 7 | A: 47-HIS; 49-GLY; 50-HIS; 75-THR; 80-ASP; 174-GLN; 196-GLN | 1 | A: 165-THR |
| 5 | Oscillamide H | 5 | A: 47-HIS; 50-HIS; 195-ASP; 232-PHE | 2 | A: 74-ASP; 135-GLN |
| 6 | Oscillamide Y | 4 | A: 195-ASP; 196-GLN; 232-PHE | - | - |
| 7 | Prenylagaramide B | 4 | A: 47-HIS; 50-HIS; 232-PHE | - | - |
| 8 | Raocyclamide A | 3 | A: 50-HIS; 84-LYS; 195-ASP | 2 | A: 74-ASP; 135-GLN |
| 9 | Anabaenopeptin B | 4 | A: 50-HIS; 195-ASP; 233-GLY | 1 | A: 73-ASP |
| 10 | Largamide A | 10 | A: 38-GLY; 40-ASP; 47-HIS; 50-HIS; 193-GLY; 195-ASP; 232-PHE | 1 | A: 73-ASP |
| 11 | Largamide B | 2 | A: 47-HIS; 196-GLN | 0 | - |
| 12 | Largamide C | 6 | A: 47-HIS; 50-HIS; 84-LYS; 195-ASP; 232-PHE | 0 | - |
| 13 | Largamide G | 6 | A: 50-HIS; 75-THR; 84-LYS; ASP-177; ASP-195; PHE-232 | 0 | - |
| 14 | 12, 13-trans-Epoxy-9-oxo-10E, 15Z-octadecadienoic acid | 2 | A: 84-LYS; 190-GLN | 1 | A: 165-THR |
| 15 | 13-Hydroperoxyoctadecadienoic acid | 1 | A: 174-GLN | 1 | A: 165-THR |
| 16 | Ergosta-8,(9)14-dien-3beta-ol | 1 | A: 224-VAL | 1 | A: 58-GLU |
| 17 | Jasmonic acid | 3 | A: 37-CYS; 38-GLY; 190-GLN | 1 | A: 165-THR |
| 18 | Methyl jasmonate | 3 | A: 37-CYS; 38-GLY; 174-GLN | 1 | A: 165-THR |
| 19 | Poriferasterol | 1 | A: 174-GLN | 1 | A: 58-GLU |
| S. No | Microalgae compound | 1JIJ | 1KZN | ||
| No of H-bond | Binding amino acid residue | No of H-bond | Binding amino acid residue | ||
| 1 | Aeruginosin 205B | 8 | A: 47-HIS; 50-HIS; 84-LYS; 193-GLY; 195-ASP; 233-GLY | 1 | A: 165-THR |
| 2 | Anabaenopeptin F | 5 | A: 47-HIS; 50-HIS; 89-VAL; 91-GLN | 5 | A: 72-GLN; 73-ASP; 135-GLN; 165-THR |
| 3 | Anabaenopeptin G | 4 | A: 47-HIS; 50-HIS; 88-ARG; 174-GLN | 2 | A: 165-THR; 73-ASP |
| 4 | Anabaenopeptin H | 7 | A: 47-HIS; 49-GLY; 50-HIS; 75-THR; 80-ASP; 174-GLN; 196-GLN | 1 | A: 165-THR |
| 5 | Oscillamide H | 5 | A: 47-HIS; 50-HIS; 195-ASP; 232-PHE | 2 | A: 74-ASP; 135-GLN |
| 6 | Oscillamide Y | 4 | A: 195-ASP; 196-GLN; 232-PHE | - | - |
| 7 | Prenylagaramide B | 4 | A: 47-HIS; 50-HIS; 232-PHE | - | - |
| 8 | Raocyclamide A | 3 | A: 50-HIS; 84-LYS; 195-ASP | 2 | A: 74-ASP; 135-GLN |
| 9 | Anabaenopeptin B | 4 | A: 50-HIS; 195-ASP; 233-GLY | 1 | A: 73-ASP |
| 10 | Largamide A | 10 | A: 38-GLY; 40-ASP; 47-HIS; 50-HIS; 193-GLY; 195-ASP; 232-PHE | 1 | A: 73-ASP |
| 11 | Largamide B | 2 | A: 47-HIS; 196-GLN | 0 | - |
| 12 | Largamide C | 6 | A: 47-HIS; 50-HIS; 84-LYS; 195-ASP; 232-PHE | 0 | - |
| 13 | Largamide G | 6 | A: 50-HIS; 75-THR; 84-LYS; ASP-177; ASP-195; PHE-232 | 0 | - |
| 14 | 12, 13-trans-Epoxy-9-oxo-10E, 15Z-octadecadienoic acid | 2 | A: 84-LYS; 190-GLN | 1 | A: 165-THR |
| 15 | 13-Hydroperoxyoctadecadienoic acid | 1 | A: 174-GLN | 1 | A: 165-THR |
| 16 | Ergosta-8,(9)14-dien-3beta-ol | 1 | A: 224-VAL | 1 | A: 58-GLU |
| 17 | Jasmonic acid | 3 | A: 37-CYS; 38-GLY; 190-GLN | 1 | A: 165-THR |
| 18 | Methyl jasmonate | 3 | A: 37-CYS; 38-GLY; 174-GLN | 1 | A: 165-THR |
| 19 | Poriferasterol | 1 | A: 174-GLN | 1 | A: 58-GLU |
Table 4: Protein-ligand interactions.
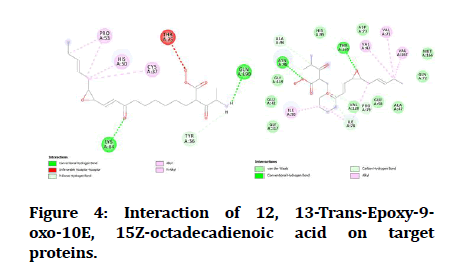
Figure 4: Interaction of 12, 13-Trans-Epoxy-9- oxo-10E, 15Z-octadecadienoic acid on target proteins.
Druglikness analysis of the selected compounds
The antagonistic interaction of inhibitors with a receptor protein or enzyme cannot guarantee the suitability of an inhibitor as a drug; therefore, drug likeness of inhibitors is more important in the drug development. Therefore all the selected compounds are screened for its drug likeness property. From analysis of 19 compounds showing higher binding energies, 6 compounds showed drug likeness. The compounds satisfying RO5 are 12,13- trans-Epoxy-9-oxo-10E,15Z-octadecadienoic acid (MW: 308 g/mol; H-bond acceptors: 4; H-bond donors: 1; MLogP: 2.26), 13-Hydroperoxyoctadecadienoic acid (MW: 312 g/mol; H-bond acceptors: 4; H-bond donors: 2; MLogP: 3.55), Ergosta-8,(9)14-dien-3beta-ol (MW: 398 g/mol; H-bond acceptors: 1; H-bond donors: 1; MLogP: 6.43), Jasmonic acid (MW: 210 g/mol; H-bond acceptors: 5; H-bond donors: 3; MLogP: 1.68), Poriferasterol(MW: 412 g/mol; H-bond acceptors: 1; H-bond donors: 1; MLogP: 6.62), Methyl jasmonate(MW: 224 g/mol; H-bond acceptors: 6; H-bond donors: 3; MLogP: 1.95).12,13- trans-Epoxy-9-oxo-10E,15Z-octadecadienoic acid, 13- Hydroperoxyoctadecadienoic acid, Jasmonic acid and Methyl jasmonate satisfied Ro5 without any violations whereas Ergosta-8,(9)14-dien-3beta-ol and Poriferasterol satisfied with 1 violation. Other compounds failed to possess drug likeness. Thus the identified 6 compounds were subjected to PASS prediction and ADMET analysis.
In silico PASS prediction
In silico PASS prediction: All nine compounds were evaluated to determine their antibacterial activity and their probable activity (Pa) and probable inactivity (Pi) values are reported. The probability of particular pharmacological action is high if Pa values are greater than Pi (Pa>Pi). Antibacterial prediction was determined for 6 compounds and presented in the Table 5. Among them 12,13-trans-Epoxy-9-oxo-10E, 15Z-octadecadienoic acid and Ergosta-8,(9)14-dien-3beta-ol showed higher Pa:Pi ratio. Notably, all the compounds showed antibacterial action i.e. Pa>Pi values.
| S. No | Microalgae compound | Pa | Pi |
|---|---|---|---|
| 1 | 12,13-trans-Epoxy-9-oxo-10E,15Z-octadecadienoic acid | 0.452 | 0.022 |
| 2 | 13-Hydroperoxyoctadecadienoic acid | 0.173 | 0.143 |
| 3 | Ergosta-8,(9)14-dien-3beta-ol | 0.315 | 0.055 |
| 4 | Jasmonic acid | 0.313 | 0.055 |
| 5 | Methyl jasmonate | 0.274 | 0.07 |
| 6 | Poriferasterol | 0.209 | 0.099 |
Table 5: PASS predictions.
ADMET properties
The ADMET characteristics of the compounds have an influence on their absorption, distribution, metabolism, excretion, and toxicity inside and throughout the human body. ADMET is a term that refers to the pharmacokinetic characteristics of a pharmacological molecule and is critical for determining its pharmaco-dynamic action. Azithromycin, a broad spectrum antibiotic is used as control drug for comparison. Azithromycin had probability values of 0.97, 0.55, 0.75, 0.67, and 0.83 for BBB, HIA, in vitro Caco-2 permeability, distribution, and non-substrate of CYP450 2C9 respectively. Other compounds showed substantial ADME values and presented in Table 6. From the toxicity analysis, AMES toxicity and mutagenicity of the compounds were evaluated. All the compounds were found to be nonmutagenic and non-carcinogenic (Table 7).
| S.No | Microalgae Compounds | In vivo blood-brain barrier penetration (C. brain/C. blood) | Human intestinal absorption (%) | In vitro Caco-2 cell permeability (nm/sec) | Distribution | CYP450 2C9 |
|---|---|---|---|---|---|---|
| 1 | Azithromycin | 0.97 | 0.55 | 0.75 | 0.67 | NS (0.83) |
| 2 | 12,13-trans-Epoxy-9-oxo-10E,15Z-octadecadienoic acid | 0.88 | 0.92 | 0.5 | 0.79 | NS (0.77) |
| 3 | 13-Hydroperoxyoctadecadienoic acid | 0.88 | 0.92 | 0.54 | 0.67 | NS (0.82) |
| 4 | Ergosta-8,(9)14-dien-3beta-ol | 0.98 | 1 | 0.83 | 0.49 | NS (0.79) |
| 5 | Jasmonic acid | 0.9 | 0.98 | 0.64 | 0.83 | NS (0.77) |
| 6 | Methyl jasmonate | 0.95 | 0.99 | 0.7 | 0.81 | NS (0.85) |
| 7 | Poriferasterol | 0.97 | 1 | 0.79 | 0.46 | NS (0.84) |
Table 6: ADME analysis.
| S.No | Microalgae Compounds | AMES Non-toxic | Non-Carcinogen |
|---|---|---|---|
| 1 | Azithromycin | 0.91 | 0.93 |
| 2 | 12,13-trans-Epoxy-9-oxo-10E,15Z-octadecadienoic acid | 0.82 | 0.88 |
| 3 | 13-Hydroperoxyoctadecadienoic acid | 0.79 | 0.67 |
| 4 | Ergosta-8,(9)14-dien-3beta-ol | 0.89 | 0.93 |
| 5 | Jasmonic acid | 0.9 | 0.88 |
| 6 | Methyl jasmonate | 0.87 | 0.84 |
| 7 | Poriferasterol | 0.91 | 0.91 |
Table 7: Toxicity analysis.
Discussion
Microalgae metabolites serve as the potential source with wide range of biological activities. In the search of new antimicrobials, microalgae metabolites can be the best solution in context of bioavailability and reactivity. Since there are thousands of metabolites identified in microalgae, in silico studies paves way for quick and effective screening of best compounds. An effective compound with target specificity and better ADMET properties can be identified. Thus, in silico screening is the best technique for identification of new drugs [13].
Today, several drug-resistant bacteria have developed with their own unique mechanisms of resistance, resulting in antibiotics that are mostly useless and a looming global threat. Antibiotic resistance (AMR) was identified by the WHO as an increasing crisis in the world, and therefore, the need for novel antimicrobial treatments to fight AMR [14]. Therefore, Microalgae metabolites from Oscillatoria sp. and Chlorella sp were screened for the antibacterial action against potential bacterial targets tyrosyl-tRNA synthetase and DNA gyrase in the present study.
Bacterial gyrase focused therapeutics is exclusive because it is the universal enzyme required for the survival of bacterial and is missing in higher eukaryotes. DNA gyrase (gyrase), a bacterial topoisomerase, is known to regulate DNA-dependent processes by introducing transient breaks in both DNA strands and relieving torsional stress in the DNA molecule by introducing negative supercoils. DNA Gyrase is a heterotetrameric protein made up of two GyrA subunits that contain the DNA cleavage site and two GyrB subunits that provide the energy for the enzyme's catalytic function by hydrolyzing ATP. Thus, drugs that target bacterial topoisomerases function in one of two ways: either by stabilising the complex between the DNA molecule and the enzyme's GyrA active site (e.g. quinolones) or by inhibiting the GyrB subunit's ATPase activity (e.g. Aminocoumarin class of inhibitors) [15].
Aminoacyl-tRNA synthetase (aaRSs) is a type of significant enzymes that catalyse the transport of amino acids to their corresponding tRNAs during protein synthesis. They are necessary to translate coded information into protein structures in nucleic acids since they recognise this information, which includes coincident tRNA molecules and amino acid structures. TyrRS are present in all living species as members of the aaRS family. TyrRS is a member of the class I tRNA synthetase family, which has two strongly symmetric sequence motifs, HIGH and KMSKS, at the active site. TyrRS in bacteria and TyrRS in humans vary in a number of ways. Small-molecule TyrRS inhibitors with these properties may be promising drug candidates for high selectivity and broad-spectrum antibacterial agents. TyrRS plays an important role in protein biosynthesis, and inhibiting these enzymes is harmful to cells. TyrRS is also highly conserved among prokaryotes, making it a good target for the production of broad-spectrum antibiotics [16].
Total of 39 compounds were identified KNApSAck database and these compounds were subjected to docking against the dual targets. Compounds showing higher binding energies (>-7 kcal/mol) were investigated for protein-ligand interaction. Interaction of ligands on binding sites of the targets proteins are very much essential on comparing with the higher binding energies. Formation of H-bonds proves the stability of the docked complex. Therefore the, H-bond formation on binding sites of the target proteins was evaluated. 14 compounds showed significant interactions with H-bonds on binding sites. Druglikness analysis was carried out based on the Lipinski rule of five (RO5). RO5 is a rule of thumb to determine if a chemical compound with a certain pharmacological or biological activity has chemical properties and physical properties that would make it a likely active drug in humans. Only six compounds showed drug likeness property and further subjected to PASS predictions and ADMET analysis. All the compounds possess Pa>Pi values for antibacterial action and had significant ADMET range on compared with Azithromycin.
Antibacterial activity of the Chlorella vulgaris and Oscillatoria geminate was investigated by Salem et al. [17]. Methanolic extracts of Chlorella vulgaris showed significant antibacterial activity against B. subtilis, K. pneumoniae, S. aureus and S. lutae; and Oscillatoria against K. pneumoniae and S. aureus. The MIC of the Oscillatoria sp. SSCM01 extracts was found to be 31.2 μg/mL against S. aureus and 7.8 μg/mL against S. typhi [1]. Similarly, Chlorella vulgaris showed MIC of 5 mg/ml against S. mutans [18]. Ethanol extracts of C. vulgaris had significant inhibition against E. coli, Klebsiella sp. and Pseudomonas sp. [19]. These results well correlates the present study.
Conclusion
Thirty-nine compounds corresponding to Oscillatoria sp. and Chlorella sp. were retrieved from KNApSAck database and used as ligands in the study. The antibacterial bacterial targets tyrosyl-tRNA synthetase and DNA Gyrase subunit b were obtained from Protein data bank. Docking analysis was performed and compounds showing higher binding energies were subjected to further Drug likeness, PASS predictions and ADMET analysis. From the analysis, we identified six compounds (12, 13-Trans-Epoxy-9-oxo-10E, 15Z-octadecadienoic acid, 13-Hydroperoxyoctadecadienoic acid, Ergosta-8, (9)14-dien-3beta-ol, Jasmonic acid, Methyl jasmonate and Poriferasterol) with potential binding energy, interactions and drug likeness properties. All the selected compounds showed significant ADME properties on compared with standard drug Azithromycin and found to be non-toxic. Thus, these compounds can be used further for development of antibacterial drugs for treatment of multidrug resistant (MDR) infections.
Acknowledgement
The author is thankful to the Deanship of Scientific Research, Prince Sattam bin Abdulaziz University, Al- Kharj, Saudi Arabia for the support in conducting the research analysis and publishing this report.
Conflict of Interest
The author declares that there are no conflicts of interest relevant to this article.
References
- Nainangu P, Antonyraj AP, Subramanian K, et al. In vitro screening of antimicrobial, antioxidant, cytotoxic activities, and characterization of bioactive substances from freshwater cyanobacteria Oscillatoria sp. SSCM01 and Phormidium sp. SSCM02. Biocatalysis Agricultural Biotechnol 2020; 29:101772.
- Elbossaty WF. Antibiotic drugs and multidrug resistance bacteria. Cit Elbossaty WF. Int J Pub Heal Safe 2017; 2:2–4.
- Rolain JM, Abat C, Jimeno MT, et al. Do we need new antibiotics?. Clin Microbiol Infection 2016; 22:408-415.
- Gangl D, Zedler JA, Rajakumar PD, et al. Biotechnological exploitation of microalgae. J Exp Botany 2015; 66:6975-90.
- Mobin SM, Chowdhury H, Alam F. Commercially important bioproducts from microalgae and their current applications–A review. Energy Procedia 2019; 160:752-60.
- https://link.springer.com/referenceworkentry/10.1007%2F978-1-4020-6754-9_13806
- Sanjay PPriya A, Shanthi S. Identification of angiotensin converting enzyme (ACE) Inhibiting phytochemical compounds from aegle marmelos, euphorbia hirta, senna auriculata, ocimum tenuiflorum and hibiscus rosasinensis by in Silic. Int J Pharma Bio Sci 2020; 11:79–85.
- Dundas J, Ouyang Z, Tseng J, et al. CASTp: Computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res 2006; 34:W116-8.
- Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Chem Biol 2015; 243-250.
- Daina A, Michielin O, Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports 2017; 7:1-3.
- Hasan M., Bhuiya N, Hossain M. In silico molecular docking, PASS prediction and ADME/T analysis for finding novel COX-2 inhibitor from Heliotropium indicum. J Complement Med Res 2019; 10:142.
- Cheng F, Li W, Zhou Y, et al. AdmetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model 2012.
- Bintener T, Pacheco MP, Sauter T. Towards the routine use of in silico screenings for drug discovery using metabolic modelling. Biochem Soc Trans 2020.
- Aslam B, Wang W, Arshad MI, et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist 2018.
- Collin F, Karkare S, Maxwell A. Exploiting bacterial DNA gyrase as a drug target: current state and perspectives. Applied Microbiol Biotechnol 2011; 92:479-97.
- Skupinska M, Stepniak P, Letowska I, et al. Natural compounds as inhibitors of tyrosyl-tRNA synthetase. Microb Drug Resistance 2017; 23:308-320.
- Salem OM, Hoballah EM, Ghazi SM, et al. Antimicrobial activity of microalgal extracts with special emphasize on Nostoc sp. Life Sci J 2014; 11:752-758.
- Jafari S, Mobasher MA, Najafipour S, et al. Antibacterial potential of Chlorella vulgaris and Dunaliella salina extracts against Streptococcus mutans. Jundishapur J Natural Pharm Products 2018; 13.
- Syed S, Arasu A, Ponnuswamy I. The uses of Chlorella Vulgaris as antimicrobial agent and as a diet: The presence of bio-active compounds which caters the vitamins, minerals in general. Int J Bio-Sci Bio-Technol 2015; 7:185–190.
Author Info
Department of Basic Medical Sciences, College of Medicine, Prince Sattam Bin Abdulaziz University, Al-Kharj, 11942, Saudi ArabiaCitation: Abubucker Peer Mohideen, Identification of Potential Metabolites in Chlorella Species and Oscillatoria Species for Treatment of Multidrug Resistant (MDR) Infections: An In Silico Analysis, J Res Med Dent Sci, 2021, 9(11): 86-94
Received: 08-Sep-2021 Accepted: 27-Oct-2021