Research - (2019) Volume 7, Issue 1
Detection of CHEK2 Ile157Thr mutation in cancer patients by using allele specific PCR
Aliarash Anoushirvani1, Azam Ahmadi2*, Mohammad Arjomandzadegan2*, Reza Aghabozorgi1, Mehrana Jafari2 and Farnaz Mehrabbeygi2
*Correspondence: Azam Ahmadi, Infectious Diseases Research Center (IDRC), Arak University of Medical Sciences, Arak, Iran, Email: Mohammad Arjomandzadegan, Infectious Diseases Research Center (IDRC), Arak University of Medical Sciences, Arak, Iran, Email:
Abstract
Aims: The checkpoint kinase 2 (CHEK2) was identified as a gene in cell cycle control upon DNA damage. The mutation in this gene is associated with some kinds of cancer including breast and lung tumors. The aim of this study was to investigate, I157T missense mutation of CHEK2 gene by Allele specific PCR (ASP) technique in clinical samples of breast and non-small cell lung cancer (NSCLC) samples.
Method: In this study, we investigated I157T mutation of CHEK2 gene in whole blood DNA from 30 breast cancers (BCs), 20 NSCLC patients and 30 control samples. The primers of ASP were designed to detect non-mutant status at the position of 430 CHEK2. The data were analyzed statistically.
Results: The results of the amplification reaction in used samples were indicated the accuracy of designed primers. The results of this study showed that in 70% of NSCLC, 33.34% of BC samples and 63.3% of control samples, the CHEK2- encoding gene contained mutations. Sequencing also confirmed these results. Statistical analysis showed that there was a direct relationship between the change in the position of 157 CHEK2 genes and metastatic NSCLC samples.
Conclusion: In previous studies, mutations at the gene coding for CHEK2 in different cancers have been proven. In the present study, it was found that the designed ASP technique was able to correctly detect the wild-type status in the studied samples and probably could be used to cancer diagnostic analysis, although more samples should be considered.
Keywords
Cancer, CHEK2 gene, ASP
Introduction
In the multistage theory of carcinogenesis, the accumulation of genetic changes leads to the formation of a tumor. Genes involved in the incidence of cancer are classified into several groups, including proto-oncogenes, tumor suppressor genes, genes involved in genome stability and cell migration. In general, many proteins involved in DNA damage repair including BRCA1,2, Her2 tyrosine kinase receptor (ErbB2), P53, PTEN, MYC, CCND1 etc. Genetic changes in each of these genes have been identified in BC and NSCLC. However, in addition to somatic and hereditary mutations, biological factors can also increase the risk for developing BC and NSCLC [1-3].
At higher levels of DNA damage, mismatch repair (MMR) pathway plays a major role in DNA repairing process and cell-cycle checkpoint control. Cell cycle regulators responding to DNA damage arrest replication and cell cycle. One of these checkpoints of the cell cycle is checkpoint kinase 2 (CHEK2) that acts during MMR repair pathway. CHEK2 plays an imperative role in response to damage or stress induction [4]. The serine/threonine kinase coded by CHEK2 is a tumor suppressor. This protein contains forkhead domains that responding to DNA damage reacts rapidly with phosphorylation and leads to inhibition of CDC25C phosphates, and ultimately prevents to mitosis which halts the cell cycle in the G1 phase. The mutation in the CHEK2 gene (Homo sapiens checkpoint kinase 2 ID: 1120) is associated with a variety of cancers, including breast, lung, colorectal, brain tumors, etc. [5]. In a study in Denmark in 2006, it was found that individuals harboring mutations in genes known as anticancer barriers including the CHEK2, p53, BRCA1, BRCA2 and ATM genes, are more sensitive to the development of cancer [6]. Bower et al. also identified a pattern of genes controlling the cell cycle associated with luminal B and basal-like cell lines [7].
Also it has been reported that Single nucleotide polymorphism (SNPs) in MMR genes can alter capacity of DNA damage in cancers and modify the function of involved genes [8]. Types of polymorphisms associated with various types of cancer, including Rs178799611, rs38416892, rs17879961, have been reported in CHEK2 gene, which have been investigated by different molecular methods. However, there are approaches for the detection of polymorphisms. In 2011, Cybulski et al. in Poland examined the risk of prostate cancer by analysis of CHEK2 gene polymorphisms using RFLP and sequencing method [9]. In molecular biology, Allele- Specific PCR technique allows efficient SNP Genotyping in a single reaction of polymerase chain reaction using thermocycler machine. In this method a reverse primer and two forward allele-specific primers amplify different allele-specific products. These fragments are detected by electrophoresis. Evaluation of different molecular methods can help to better diagnose and effectively treatment of cancer. In this study, the ATT>ACT (nucleotide number 430) mutation associated with amino acid No. 157 (rs17879961) of CHEK2 was studied in clinical samples of breast cancer and NSCLC by the allele specific method.
Materials and Methods
Sample preparation
In present case-control study, the whole blood samples were collected from Khansarinejad hospital, Arak, Iran (2017-2018). We used 50 patient’s samples with BC and NSCLC in addition 30 normal samples. DNAs were extracted using special kit (Diatom, Iran). Quality of extracted DNA was determined by 1% agarose gel (Genefanavaran, Iran) and the absorbance of A260/A280 nm in spectrophotometer (Eppendorf, Germany).
Allele specific PCR (ASP)
The sequences of designed primers for ASP technique were CHEK2-F: 5'-ATATCCAGCTCCTCTACCAG-3' and CHEK2-R: 5'-GTGATCTTCTATGTATGCAA-3'. These primers in wild-type status amplified a 375 bp fragment. We used 40 ng genomics DNA, 7.5 μl 2X master-mix (YTA, Iran) and 20 pmol of aforementioned primers in total volume 15 μl per amplification tube. This reaction was performed in thermocycler system (Eppendorf, Germany) in a touchdown protocol (annealing temperature 62°C-52°C). Results of ASP were analyzed by 1.3% agarose gel in gel doc system (Quantum ST4, Germany).
Sequencing
The confirmation of ASP method, were performed by sequencing. Amplicons were sent to Macrogene Company (South Korea) for sequencing by ABI Applied Biosystems 3730xl system.
Statistics analysis
Results were analyzed using GraphPad 7.00 and SPSS 16.0 software. Medcalc software was used for CI and Odd ratio calculation.
Results
In present study an ASP technique were designed for detection of mutation at Ile157Thr hot-spot position of CHEK2 gene. Figure 1 showed the alignment result of primers for detection of aforementioned missense mutation in used ASP.
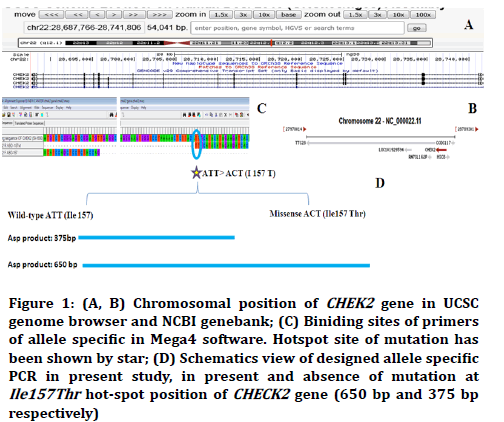
Figure 1. (A, B) Chromosomal position of CHEK2 gene in UCSC genome browser and NCBI genebank; (C) Biniding sites of primers of allele specific in Mega4 software. Hotspot site of mutation has been shown by star; (D) Schematics view of designed allele specific PCR in present study, in present and absence of mutation at Ile157Thr hot-spot position of CHECK2 gene (650 bp and 375 bp respectively)
Figure 2 showed the electrophoresis results of amplification products. These bands indicated the accuracy of the genomic DNA extraction, purification process and the PCR reaction.

Figure 2. Electrophoresis result of different patterns of allele amplification in used samples
The length of PCR Products in wild-type state was 375 bp which are related to TT genotypes. In mutant states is created a non-specific amplification at 650 bp (CC). Also 375 bp and 650 bp indicated TC genotype.
The results of ASP were validated by sequencing. The results showed the rs17879961 genotypes distributions; TT homozygote, TC heterozygote and CC mutant homozygote variant in cancer group (BC and NSCLC) were different with normal samples (Table 1).
| Genotype | BC (%) n=30 | Control (%) n= 30 | 95% CI | Odds ratio | z statistic | Significance level |
| TC heterozygote | 6.66 | 26.66 | 0.0839 to 0.4936 | 0.2035 | 3.522 | p=0.0004 |
| CC homozygote | 33.33 | 63.33 | 0.1474 to 0.4794 | 0.2658 | 4.404 | p<0.0001 |
| TT homozygote (WT) | 60 | 10 | 6.2751 to 29.0434 | 13.5 | 6.659 | p<0.0001 |
| Genotype | NSCLC n=20 | Control (%) n= 30 | 95% CI | Odds ratio | z statistic | Significancelevel |
| TC heterozygote | 0 | 26.66 | 0.0007981 to 0.2215 | 0.0133 | 3.01 | p=0.0026 |
| CC homozygote | 70 | 63.33 | 0.7599 to 2.4714 | 1.3704 | 1.047 | p=0.2950 |
| TT homozygote (WT) | 30 | 10 | 1.7666 to 8.4217 | 3.8571 | 3.388 | p=0.0007 |
| Genotype | NSCLC n=20 | BC (%) n=30 | 95% CI | Odds ratio | z statistic | Significance level |
| TC heterozygote | 0 | 6.66 | 0.003494 to 1.1011 | 0.06202 | 1.894 | p=0.0582 |
| CC homozygote | 70 | 33.33 | 2.4848 to 8.2412 | 4.5253 | 4.936 | p<0.0001 |
| TT homozygote (WT) | 30 | 60 | 1.9486 to 6.2866 | 3.5 | 4.193 | p<0.0001 |
Table 1: The different created patterns by ASP method in normal, breast cancer (BC) and non-small cell lung cancer (NSCLC) samples
Discussion
CHEK2 is one of the genes involved in various types of lung cancer, as well as classified as one of the 10 altered genes associated with hereditary BC [10-12]. In addition to, CHEK2 gene expression has been observed in colorectal, ovarian, brain and prostate cancers [13-15]. Processing of DNA damage via MMR pathway, CHEK2 nuclear protein interacts with BRCA1 and phosphorylates it and returned to survival after DNA damage. CHEK2 might be a target gene in the treatment of patients with lung cancer, however protective effect of mutant CHEK2 alleles is not yet known [16]. The ATM/ CHK2 pathway regulates the checkpoint of G1 phase in cell cycle, through activation of the p53 that prevents cells from entering S-phase [17].
Several SNPS in the encoding region of this gene have been studied in previous studies. In the present study, we developed an in-house method for detecting the mutation in the rs17879961 of CHEK2 gene, which leads to nucleotide conversion T to C and a changing in the second nucleotide of the codon 157 (isoleucin acid to threonine amino acid). The cost of used ASP method was very low and feasible in our molecular lab.
In 2016, the association between CHEK2 polymorphism and platinum-based chemotherapy efficiency in Chinese non-smokers with NSCLC was investigated and revealed that individuals with rs4035540 A/G genotype more survived [18]. In another study in 2018, a variety of polymorphisms in the CHEK2 gene, including rs1547014 and rs738722 were detected, which could possibly predict a HBV-associated hepatocellular carcinoma patient's response to therapy [19]. In a study in 2018 in the United States, CHEK2 SNPs and ATM variants were also examined [20]. In our study, we analyzed lung and BC cancer samples, however, total number with CC genotypes in lung cancer were more than BC samples (Table 1). In a 2008 study in China, the association of 1100delC 1111C>T mutation with susceptibility to BC was determined [21]. Their study showed that CHEK2 1100delC mutation may be a rare variant in Chinese populations and may not contribute to predisposition for BC. However, there may be an association between BC genetic susceptibility in China and this variant.
In our study, some of the demographic data (Table 2) of cancer patients with mutation in the amino acid 157 of the CHECK2 gene were examined and elucidated that number of metastatic and non-metastatic of NSCLC samples have alteration of 157 codon of CHEK2 gene. Although, most of the BC samples in this study were non metastatic and among them, 60% don’t have mutation in the codon 157 of CHEK2 gene.
| Characteristic-NSCLC | Detail | No. of patients (%) | Characteristic-BC | Detail | No. of patients (%) |
|---|---|---|---|---|---|
| Median age, range | ≤ 58 | 48 | Age | <40 | 23 |
| >58 | 52 | >40 | 77 | ||
| Gender | Women | 20 | Menopausal age | No | 48 |
| Men | 80 | <52 | 8 | ||
| >52 | 44 | ||||
| Smoking | Yes | 76 | BMI | >22 | 21 |
| No | 24 | <22 | 79 | ||
| Metastasis | Yes | 64 | Metastasis | Yes | 17 |
| No | 36 | No | 83 |
Table 2: Demographic data of cancer patients with mutation in the amino acid 157 of the CHECK2 gene
Luo et al., investigated the mechanism of change in the function of the CHEK2 in triple Negative BC cells, chemotherapy-resistant cell line. In their study, it was found that in the MDA-MB-231 cell lines, Y390C mutation leads to resistance to chemotherapy through the effect on the cell cycle arrest by activating the p53 and CHEK2 gene in the apoptosis process [22]. Also aforementioned mutation disrupted the normal activity of the CHEK2 gene in patients with high risk of BC. CHEK2 alteration is related to CRrispr-cas9 istop, new attractive subjects in molecular genetics also [23]. Mutations in the CHEK2 gene including I157T have been reported in the different cancers. However due to change in second nucleotide which converts isoleucine to threonine amino acids, that both of these amino acids belonged to same family with similar chemical properties.
Conclusion
In present study, we detected I157T mutation in CHECK2 gene using an ASP technique. We confirmed this alteration of CHECK2 gene is related to metastatic status of NSCLC, therefore, could be used as a marker of metastatic probably. Although the larger sample size of cancer patients will need to accurate analysis.
Acknowledgements
We thank from the office of research and technology Arak University of Medical Sciences.
Conflict of Interest
The authors declare that there is no conflict of interest regarding the publication of this manuscript.
References
- Israyelyan AH. The development of molecular diagnostics for breast cancer. LSU Master's Theses 2003.
- De Jong MM, Nolte IM, Te Meerman GJ, et al. Genes other than BRCA1 and BRCA2 involved in breast cancer susceptibility. J Med Genet 2002; 39:225-42.
- McCafferty MP, Healy NA, Kerin MJ. Breast cancer subtypes and molecular biomarkers. Diagn Histopathol 2009; 15:485-9.
- Dominguez-Valentin M, Nakken S, Tubeuf H, et al. Identification of genetic variants for clinical management of familial colorectal tumors. BMC Med Genet 2018; 19:26.
- Soyano AE, Baldeo C, Kasi PM. BRCA mutation and its association with colorectal cancer. Clin Colorectal Cancer 2018; 17:e647-50.
- Nevanlinna H, Bartek J. The CHEK2 gene and inherited breast cancer susceptibility. Oncogene 2006; 25:5912.
- Bower JJ, Vance LD, Psioda M, et al. Patterns of cell cycle checkpoint deregulation associated with intrinsic molecular subtypes of human breast cancer cells. NPJ Breast Cancer 2017; 3:9.
- Yarnold J. Early and locally advanced breast cancer: Diagnosis and treatment national institute for health and clinical excellence guideline 2009. Clin Oncol 2009; 21:159-60.
- Cybulski C, Wokołorczyk D, Jakubowska A, et al. Risk of breast cancer in women with a CHEK2 mutation with and without a family history of breast cancer. J Clin Oncol 2011; 29:3747-52.
- Kilpivaara O, Vahteristo P, Falck J, et al. CHEK2 variant I157T may be associated with increased breast cancer risk. Int J Cancer 2004; 111:543-7.
- Cybulski C, Gorski B, Huzarski T, et al. CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet 2004; 75:1131-5.
- Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell 2007; 11:103-5.
- Lawrenson K, Iversen ES, Tyrer J, et al. Common variants at the CHEK2 gene locus and risk of epithelial ovarian cancer. Carcinogenesis 2015; 36:1341-53.
- Janiszewska H, Bąk A, Pilarska M, et al. A risk of essential thrombocythemia in carriers of constitutional CHEK2 gene mutations. Haematologica 2012; 97:366-70.
- Cybulski C, Huzarski T, Górski B, et al. A novel founder CHEK2 mutation is associated with increased prostate cancer risk. Cancer Res 2004; 64:2677-9.
- Cybulski C, Masojć B, Oszutowska D, et al. Constitutional CHEK2 mutations are associated with a decreased risk of lung and laryngeal cancers. Carcinogenesis 2008; 29:762-5.
- Eymin B, Gazzeri S. Role of cell cycle regulators in lung carcinogenesis. Cell Adh Migr 2010; 4:114-23.
- Xu W, Liu D, Yang Y, et al. Association of CHEK2 polymorphisms with the efficacy of platinum-based chemotherapy for advanced non-small-cell lung cancer in Chinese never-smoking women. J Thorac Dis 2016; 8:2519.
- Wu H, Huang XP, Xiang R, et al. CHEK2 SNPs predict better prognosis in HBV-related hepatocellular carcinoma patients. Int J Clin Exp Pathol 2018; 11:3165-75.
- West AH, Blazer KR, Stoll J, et al. Clinical interpretation of pathogenic ATM and CHEK2 variants on multigene panel tests: navigating moderate risk. Fam Cancer 2018; 17:495-505.
- Chen W, Yurong S, Liansheng N. Breast cancer low-penetrance allele 1100delC in the CHEK2 gene: Not present in the Chinese familial breast cancer population. Adv Ther 2008; 25:496-501.
- Luo L, Gao W, Wang J, et al. Study on the mechanism of cell cycle checkpoint kinase 2 ( CHEK2) gene dysfunction in chemotherapeutic drug resistance of triple negative breast cancer cells. Med Sci Monit 2018; 24:3176.
- Billon P, Bryant EE, Joseph SA, et al. CRISPR-mediated base editing enables efficient disruption of eukaryotic genes through induction of STOP codons. Mol Cell 2017; 67:1068-79.
Author Info
Aliarash Anoushirvani1, Azam Ahmadi2*, Mohammad Arjomandzadegan2*, Reza Aghabozorgi1, Mehrana Jafari2 and Farnaz Mehrabbeygi2
1Khansari Hospital and Department of Internal Medicine, School of Medicine, Arak University of Medical Sciences, Arak, Iran2Infectious Diseases Research Center (IDRC), Arak University of Medical Sciences, Arak, Iran
Citation: Aliarash Anoushirvani, Azam Ahmadi, Mohammad Arjomandzadegan, Reza Aghabozorgi, Mehrana Jafari, Farnaz Mehrabbeygi, Detection of CHEK2 Ile157Thr mutation in cancer patients by using allele specific PCR , J Res Med Dent Sci, 2019, 7(1): 206-210.
Received: 29-Jan-2019 Accepted: 26-Feb-2019
