Research - (2022) Future Prospects of clinical and Medical Research
A Study on Structure and Function of Coronavirus Spike Proteins
Sonal Setya1*, Arminder Kaur2, Divya Prakash3 and Ramneet Kaur4
*Correspondence: Sonal Setya, Department of Pharmacy Practice, SGT University, Gurugram (HR), India, Email:
Abstract
Coronavirus spike enzyme, like a cell-invading biological mechanism, due of their great diversity, they provide an evolutionary challenge. This review paper discovered the cryo-EM structures of the genus' avian respiratory syncytial coronavirus spike proteins. The trimeric nϔlmmo Bronchitis Virus (IBV) spike ectodomain is made up of 3 receptors binding S1 head as well as trimeric membranes lipid S2 stalks. IBV S1 spans the gap between these different spikes and forms an evolutionary spectrum, whereas IBV S2 is structurally akin to those from new genera, and two S1 domain, comprising the N terminals domains (S1-NTD) or the C-terminals domains deviate from simple tertiaries structure or quaternaries packing to the more complicated ones across various species, resulting in varied roles of the spike in receptors use or membranes synthesis. Based on the system and functionals similarities stated above, the evolution spectra of coronavirus spike followed the sequence of γ-, δ--, and β- genera. The goal of this study is to learn more about the evolutionary relationships amongst coronavirus spike proteins while also broadening our understanding of their structural as well as functional diversity. Despite the fact that considerable study has been done in this area, there is still need for further research in the future.
Keywords
Beta Coronavirus, Membrane Fusion, Spike Protein, Syndrome
Introduction
Corona virus are a broad category of viruses that may induce a widespread ranges of illnesses, from the common colds to life-threatening disorders. A novel coronavirus is a coronavirus strain that has never been seen in humans before. Coronaviruses are dangerous to humans and other animals' health. The acute breathing distress syndromes coronaviruses infected 9,000 individuals between 2002 and 2003, with a 10% mortality rate. The Middle East breathing symptoms coronaviruses (MERS-Co Virus) has affected around 1,800 individuals since 2012, with a 36.1 percent mortality rate. Porcine epidemics diarrhea coronavirus has spread throughout United State since 2013, killing nearly all piglet or wiping obtainable more than the 1.01 percent of the country's pig herds in lesser than a year. Coronavirus causes severe breathing, gastrointestinal, or central nervous systems illnesses in people other animal, jeopardizing their health or resulting in financial losses. Coronaviruses have a remarkable ability to adapt to new settings via mutation or recombination, enabling them to change host wide range or tissue tropism quickly. Coronaviruses, as a result, constitute a long-term and continuing danger to human health [1].
The study of coronavirus virology and management of their transmission has far-reaching consequences for global health and socioeconomic stability. Corona viruses are members of the Coronaviridae family, which is a Nidovirales subfamily [2]. Alpha corona virus, Beta coronavirus, Delta corona virus, and Gamma corona virus are the four genera in which they are categorized. Coronaviruses, from alpha through beta, infect mammals, gamma or delta coronaviruses affect birds, and delta coronaviruses infect both mammalian and avian species. Coronavirus, one of the viruses that causes the common cold, has been infecting humans for a long time. Coughing, sneezing, and touching an infected surface are the main sources of infection since it is a spreadable viral diseases that might be transmitted via ingestion and inhalation of the viruses droplet. The corona virus genome is 30000 nucleotides long. As shown in Figure 1, this gene produces four proteins: nucleocapsid protein, membrane nutrients, spike protein, and envelop nutrients, as well as many non-structural proteins.
The atomic capsids, or N-protein, is linked to the viruses sole decent strand ribonucleic acids or enables it to the hijack living tissue and converts it into the viral factory. The S-protein is incorporated on the virus's surface and helps the virus enter the human host by mediating virus adherence to host cells receptor or membrane fusions between the viral & host cells membranes. The Eproteins is a small membrane of proteins with 77 to 108 amino acid that acts as a partial replacement in viral particles. It's involved in viral assembly, hosting cell membrane permeability, and virus-host cell interactions. The genetic materials are encased in a lipids sheath. The hemagglutinin-esterase dimers has been discovered on the virus-related surface. Each of the three subunits of this glycoprotein contains 1273 amino acids and two well-defined protein family regions: S1 or S2, which are involved in cell recognitions or membrane fusion, correspondingly. The latter arises as a result of a variety of yet-to-be-identified protein structural changes [3].
Figure 1: a) The genetic structure of COVID-19 and the functional domain of S protein are being shown schematically. The construction of the S protein lies underneath the genetic architecture. b) There are two subunits in the S protein: S1 and S2. A lipid bilayer surrounds the viral surfaces components spike, envelopes, or membrane [4].
Coronavirus spike protein recognize receptors
Coronaviruses recognize receptors in a convoluted way. Graph 2 both α coronavirus HCoV NL63 as well as the β coronavirus SARS-CoV identify a zinc peptidase angiotensin changing enzymes. Moreover, HCoV-NL63 as well as other alpha corona viruses target other receptors: for examples, TGEV as well as PEDV recognize a separate zinc peptidase, aminopeptidase. SARS-CoV, like other β corona viruses, recognizes dissimilar receptors: MERSCoV as well as HKU4 identify serine peptidases and a Dipeptidyl Peptidases, accordingly (DPP4). CEACAM1 (the carcinoembryonic antigens-related cells adhesion component) is a cell adhesion molecules that is recognized by the Murine Hepatitis Virus (MHV) (BCoV or OC43 detect sugar). These alpha corona viruses Transmittable Gastroenteritis Virus as Porcine Epidemic Diarrhea Virus as well as the gamma coronavirus Bronchitis virus’s spike, utilize sugar receptors and coreceptors. Many coronavirus receptors have their own physiology in addition to their involvement in viral attachment. Coronaviruses are known for their diverse receptor repertoire. S1 subunits from distinct species have relatively little structural similarities, while those out of the same genus have a lot of it (Figure 2)[5].
Figure 2: The structure of prefusion trimeric corona virus spikes was discovered using cryo–electron microscopy. (a) Spikes of the trimeric mouse hepatitis coronaviruses. 3 monomers are displayed (cyan, green or magenta). (b) Since the trimeric MHV spikes, one monomer. S1 N-terminals domains and S1 C-terminals domains, and the fusions peptides are the most essential functional components of the spikes (FP). A disordered loop is shown by the dotted curve. Two important proteolysis sites are revealed by scissors [6].
Corona viruses S1's N-terminals domains (S1-NTD) Cterminal domains have been recognized. These S1 domains may interact with receptors or serve as the receptors binding region in one or both cases. Except for the beta coronavirus MHV S1-NTD, which targets the proteins receptors CEACAM1, and the S1-NTDs are responsible for sugar binding. The proteins receptor DPP4, ACE2, ACE2, or APN is recognized by S1-CTDs. A numbers of S1 domain have had their crystal structures identified in conjunction with their receptor. Because of all these designs as well as functional studies, many of the riddles surrounding coronavirus receptor identification have been addressed [7].
Coronavirus S1-Ctds recognizes receptors
The primary atomic picture of coronaviruses S1 came from the structural of beta coronavirus. SARS-Corona Virus S1-CTD forms complexes with people ACE2, as seen in Figure 3. There are two subdomains in the SARSCorona Virus S1-CTD: the core structure or a receptorsbinding motif. And a five-stranded antiparallel sheets is the basic structure. To bind ACE2, the RBM has a slightly concave outer surface. A tiny, two-stranded antiparallel sheet with loops creating two ridges serves as the bending surface's foundation. A membranes distal peptidase domains or a membranes proximal collect in region are found in the ectodomain of ACE2. Many viruses binding motif have been discovered on the peptidase domains' outer most surface, distant from peptidase catalytic site. SARS-CoV binding has no effects on ACE2's active site, and ACE2's enzymatic activity has no effect on SARS-CoV entrance [8].
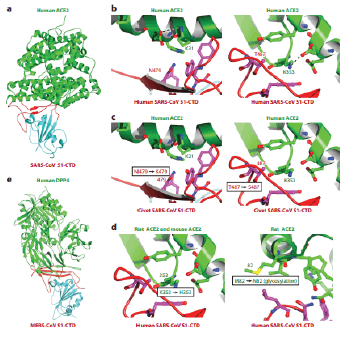
Figure 3: S1 C-terminal regions of beta coronavirus crystal structure (S1-CTDs) [8].
SARS-CoV and ACE2 interaction has provided new information on SARS-CoV cross-species spread. SARSCoV strains were discovered in both humans and palm civets via local animal markets during the SARS outbreak. And Only 2 residue in the RBM regions differ between human and civet viral S1-CTDs: Asn479.1 or Thr487.00 in human’s viral strains becomes Lys478 and Ser487 in civet’s virus strains. Human SARS-Corona Viruses S1-CTD interacts with people ACE2 considerably more close than the civet SARS-Corona Virus S1-CTD. On human ACE2, 2 virus binds hot spot have been discovered, both centered on the ACE2 residue Lys31 or Lys353, separately. And the Both hot spots are made up of a salt bond buried in a hydrophobic atmosphere, and they play very important role in virus receptors binding. SARS-CoV S1-CTD residues 479 and 488 interacts closely with the hot areas or are under the pressure to change. The hot spot designs of S1-CTD were enhanced by 2 naturally chosen viral mutation, K479N or S487T, and the finding to binding of S1-CTD for people ACE2 was increased [9].
When compared to SARS-CoV, structure of the coronavirus MERS-Corona Virus S1-CTD complex with humans DPP4 was an intriguing examples of how 2 structurally similarly viral RBDs target distinct proteins receptor. Like SARS-Corona Virus S1-CTD, MERS-Corona Virus S1-CTD has 2 subdomains: and a core structure or a RBM, Although MERS-Corona Virus or SARS-Corona Virus S1-CTDs have very similar basic architectures, their RBMs are quite differ. In contrasts to the SARS-Corona Virus RBM, which has a loops dominated and slightly concave surfaces, MERS-Corona Virus RBM has a fourstranded alternatively spliced with a moderately flat interface to binds DPP4. DPP4, on the other hands, produces a homodimers with a hydrolase region and therefore a propeller domains in each monomer. The VBMs are found on the -propeller domain's outer surface, distant from the peptidase catalytic site. Cross-species transmission of MERS-CoV is challenging owing to differences in VBM residues and DPP4 homologs across mammalian species. The VBM residues in both the mouse and rat DPP4 molecules hinder MERS-CoV binding, making them poor MERS-CoV receptors. Because of its conserved VBM residues, Camel DPP4 is an efficient MERS-CoV receptor [10].
SARS-Corona Virus S1-CTD structural complex with the humans SARS-Corona Virus S1-CTD complex with live organisms ACE2. S1-CTD's core structure (cyan), receptors binding motif, or ACE2 are all shown (green). (c) Humans beings Interfaces between SARS-Corona Virus S1-CTD or humans ACE2, including two virusbinding hot sites on humans ACE2. Salt bridge are shown by dashed lines. Humans ACE2 interacts with SARS-CoV S1-CTD in palm civets. The differences between human and civet SARS-CoV strains are shown. In living creatures, rat or mouse ACE2 interacts with SARS-CoV S1-CTD. The differences in critical residues between human, rat, and mouse ACE2 are illustrated.
Spike proteins of coronavirus trigger membrane fusion
Coronavirus spike have a more complex conformational transition trigger pattern than other class I transmembrane fusion proteins, which is likely owing to their distinctive structural features mentioned above. Despite the fact that influenza viral HA is activated by proteolysis throughout viral packing, several coronavirus spike proteins are not instead, all corona virus spike are exposed to the proteolysis advanced in the cells entry process, somewhere after receptors contact. As a consequence, coronavirus spike proteolysis may directly lead to homologous recombination, making it a key cell fusion trigger. The coronavirus spike-cleaving host proteases can be found in four stages of the virus’s infection cycle: (a) pro proteins convertases throughout viruses wrapping in the viruses producing cells, (b) the extracellular proteases afterward viruses release into the extracellular medium, (c) cells surface enzyme inhibitors after viruses connection to disease cells, (d) lysosomal proteases later virus connection to viruses infected cells. Old-style trigger including such as receptors binding or lower pH, in addition to proteolysis, might play a major roles in membrane fusion.
The results are complex and even conflicting. The MHV spikes is severed by the proprotein convertases throughout viral packaging in disease cell. For MHV to enter virus-targeting cells, this proteolysis is required. Second, CEACAM1 binding causes the MHV spikes to change conformation, resulting in membrane fusion. Incubations of MHV spikes with the recombinants solubles CEACAM1 resulted in increased hydrophobicity of the MHV S2 as well as the development of a proteases resistant S2 fragment, supporting this theory. These findings showed that fusion peptides were exposed and a six-helix bundle was formed in post-fusion S2. Third, studies differ on whether MHV penetrates target cells via endocytosis or through the cell membrane, as well as whether the MHV spike changes form at low, neutral, or even high pH.
Coronavirus spike S2 functional or structural evolution
The structure or functions of S2 from several additional coronavirus families are quite similar to IBV S2. HR2 is disordered in pre-fusion structures of IBV S2, while HR1 or FP both have many helices or linking loop ( exact residues wide ranges of FP not clear). the post fusions structures, HR1 will fold into a longer helical, HR2 into helix or coil mixture, triple copy of the HR1 or HR2 will packs into the six helix part or parcel, and FP will refolds as well as enter into the cellular membrane. Because of the structural constraint imposed by S1, IBV S2 is stuck in a pre-fusion condition. HR1 or FP of trimeric IBV S2 are organizationally limited by 2 S1-CTD from other subunits or SD1 from the another subunits, due to crosssubunit quaternary packing. Standing up S1-CTD (enabling receptor interaction) can relax structural restrictions between S1 and S2, but also S1 can be completely eliminated by proteolysis. This packing between S1 as well as S2 for IBV spiking is almost the same as co viral spikes. Although this, the packing among S1 & S2 in - and -coronavirus spike differs owing to the intra-subunits quaternary packing of each trimeric S1: HR1 and FP are limited through one S1-CTD or either SD1 from another subunits, respectively. Aside from the packing variations between S1 and S2, structural & functional similarity between coronavirus S2 from various genera indicate that co virus S2 has remained evolutionarily conserved.
Literature Review
Coronavirus sickness is a newly emerging infectious diseases that is now sweeping the world, according to Yuan Huang et al. It's affected by an original coronavirus is known as acute respiratory syndromes (SARS syndromes coronavirus 2. (SARS-Corona Virus 2). The SARS-spikes CoV-2 s (S) protein consists of two subunit, S1 and S2. The SARS-spikes Corona Virus 2 s (S) proteins is made up of two subunit: S1 as well as S2. SARS-spikes CoV-2 s (S) proteins play a critical function in receptors identified as well as cell membranes fusions. The receptors binding domains of the S1 subunit identifies or binds to the receptors of angiotensin altering enzymes, whereas the 2 heptad repeat domains of the S2 subunits enhances viral cellular membranes fusions by creating a six-helical bundles. In this review, the authors discuss recent scientific advancements in the structure, functioning, and developments of antiviral medications that target the S protein [11].
According to Berend Jan et al., the infectious spike (S) glycoprotein mediates coronavirus entry. After translation, the rat hepatitis B vaccine strain A59's 180- kDa oligomeric S protein is cleaved into a S1 ligand binding units or an S2 membranes fusion units. When the two peptides were combined, they formed a highly stable oligomeric complex. Peptides were strongly alpha helical on their own and inside the complex. The complex has a rod shape with a length of 14.5 nm, according to electron microscopy. HR1 or HR2 are antiparallel in the complex, according to limited proteolysis and mass spectrometry. The suggested fusion peptide, which is found in HR1's Nterminal domains, would be close to the transmembrane anchor in the normal protein in this shape. The HR2 peptide has been demonstrated in biological investigations to be a potent inhibitors of viral entrance into cells as well as cell-cell fusions. The coronavirus spikes proteins are a class I viral fusions proteins, according to biochemical and biophysical evidence [12].
According to Belouzard et al., the S1 domains of the coronavirus spike proteins is responsible for specific receptors, whereas the S2 domains promotes membranes fusion in early stages of viral infection. S proteins is proteolytically disrupted at the S1 or S2 border in certain instances. The endosomal proteases cathepsins L is required for viral entry by the acute breathing syndromes coronaviruses (SARS-Corona Virus); nevertheless, trypsin therapy has been demonstrated to significantly enhance SARS-Corona Virus infection. The proteolytic mechanism of a SARS-CoV S protein remained unknown in terms of the how cleavages might cause membranes fusion. Within the SARS-CoV S1 domains is the SARS-Corona Virus S2 domains (S2, R797). It was discovered that a proteolytic cleavage site exists. In both cell-cell fusion and pseudo virion entry tests, R797 mutations substantially reduced trypsin-dependent fusion. They also included a furin cleavages site at the junctions of S1 or S2, as well as the S2 cleavage site inside S2 793-KPTKR-797 (S2). Trypsin-independents cell-cell fusion was enabled by inserting a second furin cleavage site at the S1-S2 location, Our findings point to a new viral fusion protein priming mechanism including a key proteolytics cleavage event on the SARS-Corona Virus S proteins at positions 797, which works in tandem with the S1 or S2 cleavages site to promote membranes fusions or virus infectivity [13].
Humans have been infected with a diversity of coronaviruses which have being caused slight to severe breathing diseases that may lead to death, according to Guruprasad et al. Human CoVs (HCoVs) are CoV viruses that create in rodents or bats or are conveyed to human via zoonotic contact. The infection-causing fusion of viral or host cells membranes requires the binding of viral spike proteins to hosts cell receptors. Pangolins transmit SARS-Corona Virus 2, bat-borne virus (RaTG13 SARSCorona Virus) that infects humans. The occurrence of the 'PRRA sequence pattern in SARS-CoV-2 spikes proteins from humans, dogs, cats, minks, tigers, and lions suggested a comparable viral entry pathway into host cells. This research discusses the structural feature of HCoVs spikes proteins, as well as the discovery of host protein or carbohydrate receptors [14].
According to Heald-Sargent et al., coronavirus cell entry programs are distinguished by virus cells membranes combinations mediated by viral spikes proteins. Interactions with receptors, proteolysis, and acidification help Coronavirus S proteins obtain membrane fusions competence in endosomes. This page summarizes the present level of knowledge on S proteins their interaction with various entry triggers, and their responses to them. The role of receptors and proteases in viral entry is discussed, with an emphasis on type II transmembrane serine proteases (TTSPs), which have been shown to activate a variety of viral fusion protein. These or other proteases are needed for coronavirus infections, perhaps because they are located near cell-surface receptors and are therefore ready to break down incoming spikes proteins into pieces that refold to enable membrane fusions. The article ends with a discussion of how knowing how coronaviruses enter the body impacts antiviral treatment [15].
Discussion
The rapid development of viruses, especially those carrying Ribonucleic Acids (RNA), makes it difficult to their evolutionary past. Due to the viral requirements to bind many host receptor, enhance membrane-fusion effectiveness, or escape host immunes monitoring, envelope-anchored coronavirus spikes proteins are the most frequently evolving coronavirus proteins. Coronavirus spikes from four distinct families are genetically heterogeneous, and the evolutionary connections between them constitute a significant virology mystery. Because viruses must operate under specific structural and functional limitations, evolutionary knowledge about them is more likely to be discovered in their secondary structure or associated functions than in their fundamental structures. Despite intensive structural investigations of coronavirus spikes, such as X-ray crystallography or cryo-EM, the form of coronavirus spikes remains unknown, limiting a complete understanding of coronavirus spike evolutionary connections. The first cryo-EM structures of the IBV spikes ectodomain from the genus was discovered in this research, which connected the different structures of coronaviruses spike into the evolutionary spectrum or showed coronavirus spike evolutionary connections. Our research examines the morphological and functional changes in coronaviruses spike from four differ genera, demonstrating how these proteins have evolved throughout time.
To begin with, all coronaviruses S1-NTD have the same as structural fold as host galectin, suggesting that they may have evolved from them. S1 NTD have evolve from a simples galectin fold framework with just an unprotected sugar bindings sites to a partial roof on top of the original structures to a full ceiling to shield the sugar binding sites from host innate immunity (outer most layer of one coronavirus ceiling could even connect to a unique protein). The quaternary packing of S1 is affected by the partial ceiling in coronaviruses S1-NTDs, as well as the wide ceiling in -coronavirus S1-NTDs. coronavirus S1 CTD from various genera are very varied, yet they nonetheless constitute an evolutionary spectrum, with or coronaviruses S1-CTDs at the extremes or coronavirus S1 CTD in the center. S1 CTD core structures evolved from - sandwich to weaker -sandwich to -sheet, while RBM core structures evolved from tiny loops to larger loops to a longer subdomain. Viruses may be able to expand their binding site recognition as a result of RBM development, leading to S1's quaternary packing. Although the exact function of urban cornerstone development is unclear, it may permit virus to enhance receptor recognitions while also helping with S1 tertiary packing.
Conclusion
Finally, despite substantial differences in the S1 or S2 packing, the S2 of all four species is structurally preserved. In the term of structural distance from the coronavirus, quantitative structure comparisons show that Also, a phylogenetic tree was constructed consuming the amino acids of corona viruses spike from several genera, with the results showing that coronavirus spikes is the nearly, coronavirus spikes is the farthest, or coronavirus spikes is intermediates S6 in term of amino acids sequence. When coronavirus spikes from various genera are combined, they form an original spectrum with the coronaviruses spike on the ones ends and coronaviruses spikes in the middle. Viruses are ideal model species for understanding evolution because of their rapid evolutionary rates. Despite structural variations across coronaviruses spikes, especially in their S1 where there are few or no structural similarities, our study has shown that thorough examination of their structure and activity may still be utilized to track phylogenetic changes among specific viral proteins. Our findings also suggest that coronaviruses spikes have grown to a remarkable level of variety in order to broaden receptors recognition, enhance membranes fusion, and elude host immunes defenses. Surveillance though the preserving basic membranes fusions mechanism. And the goal of this study is to provide light on the evolutionary relationships amongst coronavirus spike proteins while also broadening understands of their structural or functional variety. Despite the fact that much study has been done in this area, there is still a lot much to be done in future.
References
- Guruprasad, L. (2020). Evolutionary relationships and sequence-structure determinants in human SARS coronavirus-2 spike proteins for host receptor recognition. ChemRxiv.
[Crossref], [GoogleScholar], [Indexed]
- Shang, J., Zheng, Y., Yang, Y., Liu, C., Geng, Q., Luo, C., Zhang, W., & Li, F. (2018). Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins. PLOSPathogens, 14(4), e1007009.
[Crossref], [GoogleScholar], [Indexed]
- Chu, H., Chan, C. M., Zhang, X., Wang, Y., Yuan, S., Zhou, J., Au-Yeung, R. K., Sze, K. H., Yang, D., Shuai, H., Hou, Y., Li, C., Zhao, X., Poon, V. K., Leung, S. P., Yeung, M. L., Yan, J., Lu, G., Jin, D. Y., .â?¯.â?¯. Yuen, K. Y. (2018). Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. Journal of BiologicalChemistry, 293(30), 11709â??11726.
[Crossref], [GoogleScholar], [Indexed]
- Kirchdoerfer, R. N., Cottrell, C. A., Wang, N., Pallesen, J., Yassine, H. M., Turner, H. L., Corbett, K. S., Graham, B. S., McLellan, J. S., & Ward, A. B. (2016). Pre-fusion structure of a human coronavirus spike protein. Nature, 531(7592), 118â??121.
[Crossref], [GoogleScholar], [Indexed]
- Evolutionary relationships and sequence-structure determinants in human SARS coronavirus-2 spike proteins for host receptor recognition
- Pillay, T. S., & Pillay, T. S. (2020). Gene of the month: The 2019-nCoV/SARS-CoV-2 novel coronavirus spike protein. Journal of ClinicalPathology, 73(7), 366â??369.
[Crossref], [GoogleScholar], [Indexed]
- de Andrade, J., Gonçalves, P. F. B., & Netz, P. A. (2021). Why does the novel coronavirus spike protein interact so strongly with the human ACE2? A thermodynamic answer. ChemBioChem, 22(5), 865â??875.
[Crossref], [GoogleScholar], [Indexed]
- Li, F. (2016). Structure, function, and evolution of coronavirus spike proteins. Annual Review of Virology, 3(1), 237â??261.
[Crossref], [GoogleScholar], [Indexed]
- Jaimes, J. A., Millet, J. K., Goldstein, M. E., Whittaker, G. R., & Straus, M. R. (2019). A fluorogenic peptide cleavage assay to screen for proteolytic activity: Applications for coronavirus spike protein activation. Journal of VisualizedExperiments: JoVE, (143).
[Crossref], [GoogleScholar], [Indexed]
- Shang, J., Wan, Y., Liu, C., Yount, B., Gully, K., Yang, Y., Auerbach, A., Peng, G., Baric, R., & Li, F. (2020). Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry. PLOSPathogens, 16(3), e1008392.
[Crossref], [GoogleScholar], [Indexed]
- Huang, Y., Yang, C., Xu, X. F., Xu, W., & Liu, S. W. (2020). Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacologica Sinica, 41(9), 1141â??1149.
[Crossref], [GoogleScholar], [Indexed]
- Bosch, B. J., van der Zee, R., de Haan, C. A. M., & Rottier, P. J. M. (2003). The coronavirus spike protein is a Class I virus fusion protein: Structural and functional characterization of the fusion core complex. Journal of Virology, 77(16), 8801â??8811.
[Crossref], [GoogleScholar], [Indexed]
- Belouzard, S., Millet, J. K., Licitra, B. N., & Whittaker, G. R. (2012). Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses, 4(6), 1011â??1033.
[Crossref], [GoogleScholar], [Indexed]
- Guruprasad, L. (2021). Human coronavirus spike protein-host receptor recognition. Progress in Biophysics and Molecular Biology, 161, 39â??53.
[Crossref], [GoogleScholar], [Indexed]
- Heald-Sargent, T., & Gallagher, T. (2012). Ready, set, fuse! the coronavirus spike protein and acquisition of fusion competence. Viruses, 4(4), 557â??580.
[Crossref], [GoogleScholar], [Indexed]
Author Info
Sonal Setya1*, Arminder Kaur2, Divya Prakash3 and Ramneet Kaur4
1Department of Pharmacy Practice, SGT University, Gurugram (HR), India2Department of Biotechnology, Sanskriti University, Mathura, Uttar Pradesh, India
3School of Biotechnology & Bioinformatics, Shobhit Institute of Engineering and Technology (Deemed to, India
4Department of Biotechnology, RIMT University, Mandi Gobindgarh, Punjab, India
Received: 02-May-2022, Manuscript No. JRMDS-22-58459;; , Pre QC No. JRMDS-22-58459(PQ); Editor assigned: 04-May-2022, Pre QC No. JRMDS-22-58459(PQ); Reviewed: 14-May-2022, QC No. JRMDS-22-58459; Revised: 18-May-2022, Manuscript No. JRMDS-22-58459(R); Published: 02-Jun-2022


